
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 26,155,846 – 26,155,957 |
| Length | 111 |
| Max. P | 0.704597 |

| Location | 26,155,846 – 26,155,957 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 79.44 |
| Mean single sequence MFE | -26.28 |
| Consensus MFE | -11.68 |
| Energy contribution | -12.57 |
| Covariance contribution | 0.89 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.42 |
| Structure conservation index | 0.44 |
| SVM decision value | 0.24 |
| SVM RNA-class probability | 0.649051 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
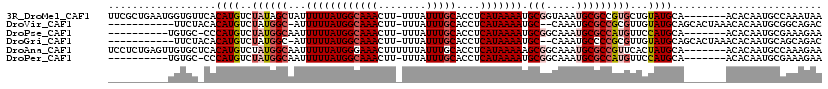
>3R_DroMel_CAF1 26155846 111 + 27905053 UUCGCUGAAUGGUGUUCACAUGUCUAUAGCUAUUUUUAUGGCAAACUU-UUUAUUUGCACCUCAUAAAAUGCGGUAAAUGCGCCGUGCUGUAUGCA-------ACACAAUGCCAAAUAA .........(((((((....(((.((((((...((((((((((((...-....)))))....))))))).(((((......))))))))))).)))-------....)))))))..... ( -26.90) >DroVir_CAF1 196444 104 + 1 -----------UUCUACACAUGUCUAUGGC-AUUUUUAUGGCAAACUU-UUUAUUUGCACCUCAUAAAAUGC--CAAAUGCGCCGCGUUGUAUGCAGCACUAAACACAAUGCGGCAGAC -----------..........((((.((((-(((((.((((((((...-....)))))....))))))))))--)).....((((((((((..............)))))))))))))) ( -32.44) >DroPse_CAF1 148710 100 + 1 ----------UGUGC-CCCAUGUCUAUGGCAAUUUUUAUGGCAAACUU-UUUAUUUGCACCUCAUAAAAUGCGGCAAAUGCGCCAUGUUCCAUGCA-------ACACAAUGCGAAAGAA ----------((((.-..((((..((((((...((((((((((((...-....)))))....))))))).(((.....)))))))))...))))..-------.))))...(....).. ( -23.40) >DroGri_CAF1 182243 104 + 1 -----------UUCUACACAUGUCUAUGGC-AUUUUUAUGGCAAACUU-UUUAUUUGCACCUCAUAAAAUGC--CAAAUGCCCCGCGUUGUAUGCAGCACUAAACACAAUGCAGCAGAC -----------..........((((.((((-(((((.((((((((...-....)))))....))))))))))--))...((...(((((((..............))))))).)))))) ( -23.74) >DroAna_CAF1 150010 112 + 1 UCCUCUGAGUUGUGCUCACAUGUCUAUGGCAAUUUUUAUGGGAAACUUUUUUAUUUGCACCUCAUAAAAAGCGGCAAAUGCGCCGUUCACUAUGCA-------ACACAAUGCCAAAGAA .....((((.....))))....(((.(((((.((((((((((..................))))))))))(((((......)))))..........-------......))))).))). ( -27.77) >DroPer_CAF1 151069 100 + 1 ----------UGUGC-CCCAUGUCUAUGGCAAUUUUUAUGGCAAACUU-UUUAUUUGCACCUCAUAAAAUGCGGCAAAUGCGCCAUGUUCCAUGCA-------ACACAAUGCGAAAGAA ----------((((.-..((((..((((((...((((((((((((...-....)))))....))))))).(((.....)))))))))...))))..-------.))))...(....).. ( -23.40) >consensus __________UGUGC_CACAUGUCUAUGGCAAUUUUUAUGGCAAACUU_UUUAUUUGCACCUCAUAAAAUGCGGCAAAUGCGCCGUGUUCUAUGCA_______ACACAAUGCGAAAGAA ..................((((..((((((...((((((((((((........)))))....))))))).(((.....)))))))))...))))......................... (-11.68 = -12.57 + 0.89)

| Location | 26,155,846 – 26,155,957 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 79.44 |
| Mean single sequence MFE | -30.40 |
| Consensus MFE | -13.71 |
| Energy contribution | -14.02 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.46 |
| Structure conservation index | 0.45 |
| SVM decision value | 0.36 |
| SVM RNA-class probability | 0.704597 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
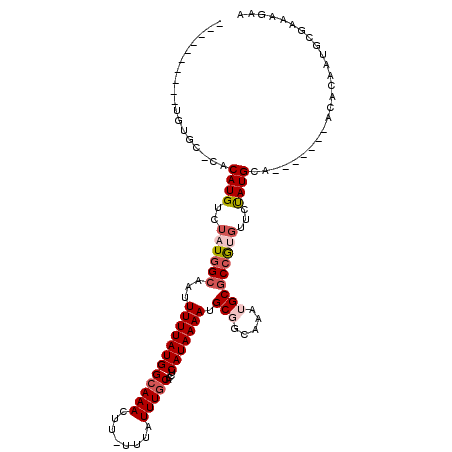
>3R_DroMel_CAF1 26155846 111 - 27905053 UUAUUUGGCAUUGUGU-------UGCAUACAGCACGGCGCAUUUACCGCAUUUUAUGAGGUGCAAAUAAA-AAGUUUGCCAUAAAAAUAGCUAUAGACAUGUGAACACCAUUCAGCGAA .......((.((((((-------((....)))))))).))......(((.(((((((....((((((...-..)))))))))))))...............((((.....))))))).. ( -23.60) >DroVir_CAF1 196444 104 - 1 GUCUGCCGCAUUGUGUUUAGUGCUGCAUACAACGCGGCGCAUUUG--GCAUUUUAUGAGGUGCAAAUAAA-AAGUUUGCCAUAAAAAU-GCCAUAGACAUGUGUAGAA----------- .(((((.(((...(((((((((((((.......)))))))...((--((((((((((....((((((...-..))))))))).)))))-)))))))))))))))))).----------- ( -37.40) >DroPse_CAF1 148710 100 - 1 UUCUUUCGCAUUGUGU-------UGCAUGGAACAUGGCGCAUUUGCCGCAUUUUAUGAGGUGCAAAUAAA-AAGUUUGCCAUAAAAAUUGCCAUAGACAUGGG-GCACA---------- ...........(((((-------(.((((....(((((((.......)).(((((((....((((((...-..)))))))))))))...)))))...)))).)-)))))---------- ( -28.90) >DroGri_CAF1 182243 104 - 1 GUCUGCUGCAUUGUGUUUAGUGCUGCAUACAACGCGGGGCAUUUG--GCAUUUUAUGAGGUGCAAAUAAA-AAGUUUGCCAUAAAAAU-GCCAUAGACAUGUGUAGAA----------- .....((((((.(((((((((.((((.......)))).))...((--((((((((((....((((((...-..))))))))).)))))-)))))))))))))))))..----------- ( -34.80) >DroAna_CAF1 150010 112 - 1 UUCUUUGGCAUUGUGU-------UGCAUAGUGAACGGCGCAUUUGCCGCUUUUUAUGAGGUGCAAAUAAAAAAGUUUCCCAUAAAAAUUGCCAUAGACAUGUGAGCACAACUCAGAGGA ((((((((..((((((-------(.((((((...(((((....))))).((((((((.((.((..........))..)))))))))).........)).))))))))))).)))))))) ( -28.80) >DroPer_CAF1 151069 100 - 1 UUCUUUCGCAUUGUGU-------UGCAUGGAACAUGGCGCAUUUGCCGCAUUUUAUGAGGUGCAAAUAAA-AAGUUUGCCAUAAAAAUUGCCAUAGACAUGGG-GCACA---------- ...........(((((-------(.((((....(((((((.......)).(((((((....((((((...-..)))))))))))))...)))))...)))).)-)))))---------- ( -28.90) >consensus UUCUUUCGCAUUGUGU_______UGCAUACAACACGGCGCAUUUGCCGCAUUUUAUGAGGUGCAAAUAAA_AAGUUUGCCAUAAAAAUUGCCAUAGACAUGUG_ACACA__________ .......(((((((((........)))))......(((((.......)).(((((((....((((((......)))))))))))))...)))......))))................. (-13.71 = -14.02 + 0.31)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:43:13 2006