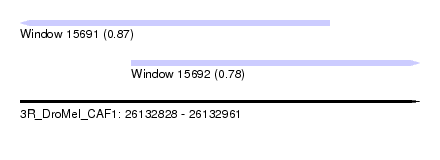
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 26,132,828 – 26,132,961 |
| Length | 133 |
| Max. P | 0.870244 |

| Location | 26,132,828 – 26,132,931 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.18 |
| Mean single sequence MFE | -34.77 |
| Consensus MFE | -18.20 |
| Energy contribution | -19.09 |
| Covariance contribution | 0.89 |
| Combinations/Pair | 1.22 |
| Mean z-score | -2.63 |
| Structure conservation index | 0.52 |
| SVM decision value | 0.87 |
| SVM RNA-class probability | 0.870244 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 26132828 103 - 27905053 UCCUGUCAGCUCCAGCUAUAU--------------AGCUUAUAGCAUAUAGCUGUCAUUGGAACAAUCAAUGGGCAUUGGAUUUUGG-GGGCUGGCCG--GUUCUUCCUGUCUGAAAUUU .((.(((((((((((((((((--------------.((.....)))))))))).(((((((.....))))))).............)-)))))))).)--)................... ( -33.80) >DroSec_CAF1 123400 100 - 1 UCCUGUCAGCUCCAGCUAUAU--------------AGCUUAUAGCAUAUAGCUGUCAUUGGAACAAUCAAUGGGCAUUGGAUUU-CU-GGGCUGGCCG--GUUCUUCCUGUCUGGAAU-- .((.(((((((((((((((((--------------.((.....)))))))))))(((((((.....)))))))...........-..-)))))))).)--)...((((.....)))).-- ( -35.30) >DroSim_CAF1 129297 99 - 1 UCCUGUCAGCUCCAGCUAUAU--------------AGCUUAUAGCAUAUAGCUGUCAUUGGAACAAUCAAUGGGCAUUGGAUU--GG-GGGCUGGCCG--GUUCUUCCUGUCUGGAAU-- .((.(((((((((((((((((--------------.((.....))))))))))..........((((((((....))).))))--))-)))))))).)--)...((((.....)))).-- ( -37.30) >DroEre_CAF1 132089 115 - 1 UCCUGUCAGCUCCAGCUAUAUUGCUUAUAGCUUAUAGCUUAUAGCAUAUAGCUGUCAUUGGAACAAUCAAUGGGCAUUGGAUUU-GG-GGGCUGGCCG--GUUCUUCCUGUCUGGAAUU- .((.(((((((((((((((((.(((...(((.....)))...))))))))))).(((((((.....)))))))...........-.)-)))))))).)--)...((((.....))))..- ( -42.60) >DroYak_CAF1 130853 111 - 1 UCCUGUCAGCUUCAGCUAUAUAGCUUAU-----AUAGCUUAUAGCAUAUAGCUGUCAUUGGAACAAUCAAGGGGCAUUCGAUUU-GGGGGGCUGGCCG--GUUCUUCCUGUCUGGAAUU- .((.(((((((((((((((((.(((.((-----(.....))))))))))))))(((.((((.....))))..))).........-..))))))))).)--)...((((.....))))..- ( -34.90) >DroPer_CAF1 133585 108 - 1 UCCUGUCAAUCCC--CUAGACCGCCC----CCCACCCCCUUUGGCCGAUAGCUGUCAUUGGAACAAUCAGUCAGCAUUCGAUU--GG-UCCGUGGUCAGUGUUCUUCCUGUCCGAC-U-- ....(((......--...(((((((.----............)))(((..((((..(((((.....)))))))))..)))...--))-))...((.(((........))).)))))-.-- ( -24.72) >consensus UCCUGUCAGCUCCAGCUAUAU______________AGCUUAUAGCAUAUAGCUGUCAUUGGAACAAUCAAUGGGCAUUGGAUUU_GG_GGGCUGGCCG__GUUCUUCCUGUCUGGAAU__ ....((((((((..((((((...................)))))).....(((..((((((.....))))))))).............))))))))........((((.....))))... (-18.20 = -19.09 + 0.89)

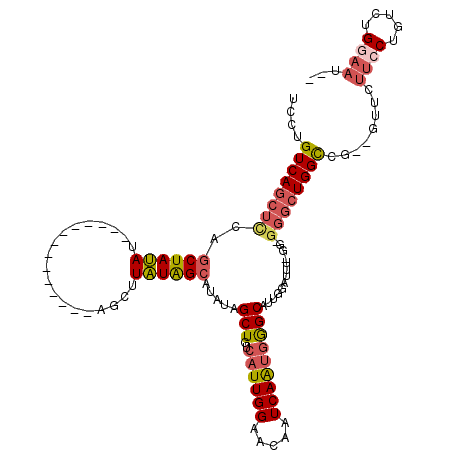
| Location | 26,132,865 – 26,132,961 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 79.80 |
| Mean single sequence MFE | -29.68 |
| Consensus MFE | -16.07 |
| Energy contribution | -16.24 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.29 |
| Mean z-score | -2.23 |
| Structure conservation index | 0.54 |
| SVM decision value | 0.56 |
| SVM RNA-class probability | 0.782886 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 26132865 96 + 27905053 CCAAUGCCCAUUGAUUGUUCCAAUGACAGCUAUAUGCUAUAAGCU--------------AUAUAGCUGGAGCUGACAGGAGGCGGAUCU--GACUCAUGUAGAUCUCGAAAU--- .....(((..(((...((((((......(((((((((.....)).--------------)))))))))))))...)))..)))((((((--(.(....))))))))......--- ( -29.30) >DroSec_CAF1 123434 94 + 1 CCAAUGCCCAUUGAUUGUUCCAAUGACAGCUAUAUGCUAUAAGCU--------------AUAUAGCUGGAGCUGACAGGAGGCGGAUCU--GACUCAU--AUAUUUCGAAAU--- ....((((..(((...((((((......(((((((((.....)).--------------)))))))))))))...)))..)))).....--.......--............--- ( -23.50) >DroSim_CAF1 129330 94 + 1 CCAAUGCCCAUUGAUUGUUCCAAUGACAGCUAUAUGCUAUAAGCU--------------AUAUAGCUGGAGCUGACAGGAGGCGGAUCU--GACUCAU--AGAUUUCGAAAU--- .....(((..(((...((((((......(((((((((.....)).--------------)))))))))))))...)))..)))((((((--(.....)--))))))......--- ( -25.70) >DroEre_CAF1 132124 108 + 1 CCAAUGCCCAUUGAUUGUUCCAAUGACAGCUAUAUGCUAUAAGCUAUAAGCUAUAAGCAAUAUAGCUGGAGCUGACAGGAGGCGGAUCU--GACUCAU--AGAUCUCGAAAC--- .....(((..(((...((((((......((((((((((...(((.....)))...))).)))))))))))))...)))..)))((((((--(.....)--))))))......--- ( -32.60) >DroYak_CAF1 130889 103 + 1 CGAAUGCCCCUUGAUUGUUCCAAUGACAGCUAUAUGCUAUAAGCUAU-----AUAAGCUAUAUAGCUGAAGCUGACAGGAGGCGGAUCU--GACUCAU--AGAUCUCGAAAC--- (((.((((.((((((((...))))..(((((.((.((((((((((..-----...)))).)))))))).))))).)))).))))(((((--(.....)--))))))))....--- ( -33.00) >DroPer_CAF1 133619 105 + 1 CGAAUGCUGACUGAUUGUUCCAAUGACAGCUAUCGGCCAAAGGGGGUGGG----GGGCGGUCUAG--GGGAUUGACAGGAGGCGGAUGUGUGACUCGC--GAAUC--GAAAAAGC (((.((((..(((...(((((...(((.(((..(.(((......))).).----.))).)))...--)))))...)))..))))..((((.....)))--)..))--)....... ( -34.00) >consensus CCAAUGCCCAUUGAUUGUUCCAAUGACAGCUAUAUGCUAUAAGCU______________AUAUAGCUGGAGCUGACAGGAGGCGGAUCU__GACUCAU__AGAUCUCGAAAU___ ....((((..(((...(((((......(((((((((((...)))...............)))))))))))))...)))..))))(((((...........))))).......... (-16.07 = -16.24 + 0.17)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:43:03 2006