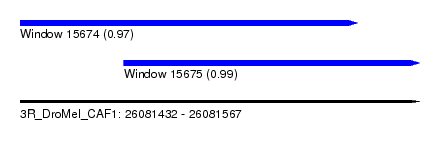
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 26,081,432 – 26,081,567 |
| Length | 135 |
| Max. P | 0.992161 |

| Location | 26,081,432 – 26,081,546 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 93.50 |
| Mean single sequence MFE | -33.18 |
| Consensus MFE | -27.98 |
| Energy contribution | -27.98 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.84 |
| SVM decision value | 1.59 |
| SVM RNA-class probability | 0.966174 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 26081432 114 + 27905053 UCUCAAGAUCAAGAGUGAACCACCCUGCUUUUUGUGGCAAGGGGCACGCACAUGCAUCAUGCAACAUGCAACAGCGGCGUGAUUGCAAUGCAGCGAGCAAAAAAA-AAAGCUGAA .(((........))).......(((((((......))).))))((.(((...(((((..((((((((((.......))))).))))))))))))).)).......-......... ( -33.20) >DroSec_CAF1 72908 108 + 1 CUUCAAGAUCAAGAGUGAACCACCCUGCUUUUCGAGGCAAGCGGCACGCACAUGCAUCAUGCAACAUGCAACAGCGGCGUGAUUGCAAUGCAGCGA------AAA-AAAGCUGGA .......................((.((((((((..(((.(((((((((...(((.....)))...(((....)))))))).))))..)))..)))------...-))))).)). ( -30.00) >DroSim_CAF1 74568 108 + 1 UCUCAAGAUCAAGAGUGAGCCACCCUGCUUUUCGAGGCAAGCGGCACGCACAUGCAUCAUGCAACAUGCAACAGCGGCGUGAUUGCAAUGCAGCGA------AAA-AAAGCUGGA ......(((((.(.(((.(((.(..(((((....))))).).))).))).).)).))).((((((((((.......))))).)))))...((((..------...-...)))).. ( -32.80) >DroEre_CAF1 77873 109 + 1 UCUCAAGAUCAAGAGUGAACCACCCUGCUUUUCGUGGCAAGGGGCACGCACAUGCAUCAUGCAACAUGCAACAGCGGCGUGAUUGCAAUGCAGCGA------AAAAAAAGCUGAA ......(((((.(.(((..((.((.((((......)))).))))..))).).)).))).((((((((((.......))))).)))))...((((..------.......)))).. ( -31.30) >DroYak_CAF1 77574 108 + 1 UCCCAAGAGCAAGAGUGAGCCACCCUGCUUUUCGAGGCAAGGGGCACGCACAUGCAUCAUGCAACAUGCAACAGCGGCGUGAUUGCAAUGCAGCGA------AAA-AAAGCUGGA ......(((((.(.(((.(((.((.(((((....))))).))))).))).).))).)).((((((((((.......))))).)))))...((((..------...-...)))).. ( -38.60) >consensus UCUCAAGAUCAAGAGUGAACCACCCUGCUUUUCGAGGCAAGGGGCACGCACAUGCAUCAUGCAACAUGCAACAGCGGCGUGAUUGCAAUGCAGCGA______AAA_AAAGCUGGA ..(((.........(((.....(((((((......))).)))).....))).(((((..((((((((((.......))))).))))))))))...................))). (-27.98 = -27.98 + 0.00)

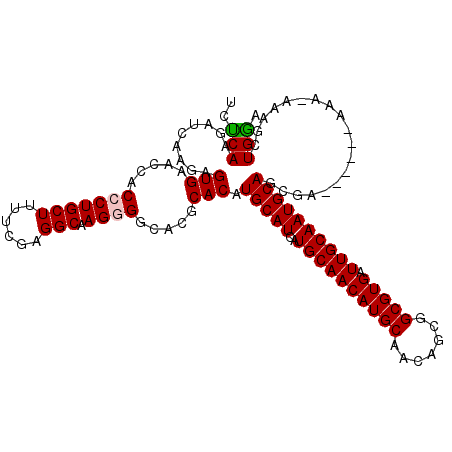
| Location | 26,081,467 – 26,081,567 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 101 |
| Reading direction | forward |
| Mean pairwise identity | 92.94 |
| Mean single sequence MFE | -28.86 |
| Consensus MFE | -25.58 |
| Energy contribution | -25.74 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.40 |
| Structure conservation index | 0.89 |
| SVM decision value | 2.31 |
| SVM RNA-class probability | 0.992161 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 26081467 100 + 27905053 GGCAAGGGGCACGCACAUGCAUCAUGCAACAUGCAACAGCGGCGUGAUUGCAAUGCAGCGAGCAAAAAAA-AAAGCUGAAAAUGCAUCGAAGUGGCAUCUA ....((((.(((....((((((..((((((((((.......))))).)))))...((((...........-...))))...))))))....))).).))). ( -27.54) >DroSec_CAF1 72943 94 + 1 GGCAAGCGGCACGCACAUGCAUCAUGCAACAUGCAACAGCGGCGUGAUUGCAAUGCAGCGA------AAA-AAAGCUGGAAAUGCAUCGAAGUGGCAUCUA .......(.(((....((((((..((((((((((.......))))).))))).(.((((..------...-...)))).).))))))....))).)..... ( -28.70) >DroSim_CAF1 74603 94 + 1 GGCAAGCGGCACGCACAUGCAUCAUGCAACAUGCAACAGCGGCGUGAUUGCAAUGCAGCGA------AAA-AAAGCUGGAAAUGCAUCGAAGUGGCAUCUA .......(.(((....((((((..((((((((((.......))))).))))).(.((((..------...-...)))).).))))))....))).)..... ( -28.70) >DroEre_CAF1 77908 95 + 1 GGCAAGGGGCACGCACAUGCAUCAUGCAACAUGCAACAGCGGCGUGAUUGCAAUGCAGCGA------AAAAAAAGCUGAAAAUGCAUCGGAGUGGCAUCUA ....((..((.(((.(((((((..((((((((((.......))))).)))))...((((..------.......))))...)))))).)..)))))..)). ( -28.60) >DroYak_CAF1 77609 94 + 1 GGCAAGGGGCACGCACAUGCAUCAUGCAACAUGCAACAGCGGCGUGAUUGCAAUGCAGCGA------AAA-AAAGCUGGAAAUGCAUCGAAGUGGCAUCUA ....((((.(((....((((((..((((((((((.......))))).))))).(.((((..------...-...)))).).))))))....))).).))). ( -29.10) >DroAna_CAF1 82333 94 + 1 UUCAAGAGGCACGCACAUGCAUCAUGCAACAUGCAACAGCGGCGUGAUUGCAACGCAGCAG------AAA-AAAACUGCAUAUGCAUCGAUGUGGCAUCUA ....(((.((.(((((((((((..((((((((((.......))))).))))).....((((------...-....))))..)))))).).)))))).))). ( -30.50) >consensus GGCAAGGGGCACGCACAUGCAUCAUGCAACAUGCAACAGCGGCGUGAUUGCAAUGCAGCGA______AAA_AAAGCUGGAAAUGCAUCGAAGUGGCAUCUA .......(.(((....((((((..((((((((((.......))))).)))))...((((...............))))...))))))....))).)..... (-25.58 = -25.74 + 0.17)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:42:48 2006