
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 3,162,617 – 3,162,771 |
| Length | 154 |
| Max. P | 0.996056 |

| Location | 3,162,617 – 3,162,735 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.12 |
| Mean single sequence MFE | -25.66 |
| Consensus MFE | -24.22 |
| Energy contribution | -24.10 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.10 |
| Structure conservation index | 0.94 |
| SVM decision value | 0.17 |
| SVM RNA-class probability | 0.616025 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 3162617 118 - 27905053 GGCUGCCGGUAAUGGAGACCAGCCCGAAAGUACAUGCACCAGUAAGAGUAUUUUUCAGACGAAAAUAUUUUUGCAUAAAUUACAAUAACAGUGGCAGAACAAGAGG--AACAACAAUGAA ..((((((((...(....)..(..(....)..)....))).((((((((((((((.....))))))))))))))..................))))).........--............ ( -25.90) >DroSec_CAF1 10503 118 - 1 GGCUGCCGGUAAUGGAGACUAGCCCGAAAGUACAUGCACCAGUAAGAGUAUUUUUCAGACGAAAAUAUUUUUGCAUAAAUUACAAUAACAGUGGCAGAACAAGAGG--AACAACAAUGAA ..((((((((.(((...(((........))).)))..))).((((((((((((((.....))))))))))))))..................))))).........--............ ( -27.00) >DroSim_CAF1 12672 118 - 1 GGCUGCCGGUAAUGGAGACCAGCCCGAAAGUACAUGCACCAGUAAGAGUAUUUUUCAGACGAAAAUAUUUUUGCAUAAAUUACAAUAACAGUGGCAGAACACGAGG--AACAACAAUGAA ..((((((((...(....)..(..(....)..)....))).((((((((((((((.....))))))))))))))..................))))).........--............ ( -25.90) >DroEre_CAF1 2190 119 - 1 GGCUGCCGGUAAUGGAGACCA-CCGGAAAGUACAUGCACCAGUGAGAGUAUUUUUCAGACGAAGAUAUUUUUGCAUAAAUUACUAUAGCAGUGGCAGAACAAGAGGCAAACAACAAUGAA .(((.(((((...(....).)-))))...((...(((....(..(((((((((((.....)))))))))))..)......((((.....)))))))..))....)))............. ( -27.30) >DroYak_CAF1 11933 117 - 1 GGCUGCCGGUAAUGGAGACCGGCCCGAAAGUACAUGCACCAGUAAGAGUAUUUUUCAGACGAAAAUAUU-UUGCAUAAAUUACAAUAACAGUGGCAGAAGACGGGG--AACAACAAUGAA ((..((((((.......))))))))....((...(((....((((((....(((((....)))))..))-))))......(((.......))))))....))....--............ ( -22.20) >consensus GGCUGCCGGUAAUGGAGACCAGCCCGAAAGUACAUGCACCAGUAAGAGUAUUUUUCAGACGAAAAUAUUUUUGCAUAAAUUACAAUAACAGUGGCAGAACAAGAGG__AACAACAAUGAA ..((((((((.(((...((..........)).)))..))).((((((((((((((.....))))))))))))))..................)))))....................... (-24.22 = -24.10 + -0.12)

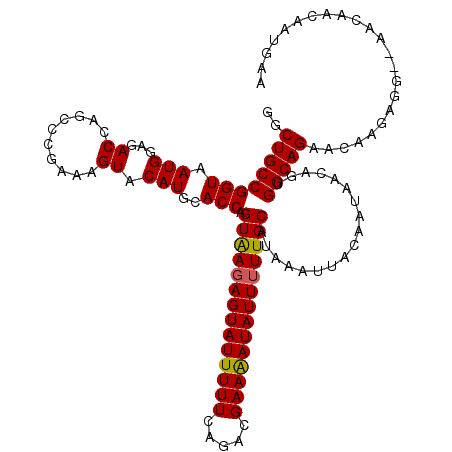

| Location | 3,162,655 – 3,162,771 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.13 |
| Mean single sequence MFE | -36.06 |
| Consensus MFE | -32.90 |
| Energy contribution | -32.58 |
| Covariance contribution | -0.32 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.90 |
| Structure conservation index | 0.91 |
| SVM decision value | 2.65 |
| SVM RNA-class probability | 0.996056 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 3162655 116 - 27905053 GGCGGUC----GCAUCUCCGACAUCUGGAGAAGAAGCCAAGGCUGCCGGUAAUGGAGACCAGCCCGAAAGUACAUGCACCAGUAAGAGUAUUUUUCAGACGAAAAUAUUUUUGCAUAAAU ...(((.----(((((((((....((((......(((....))).))))...))))))...(..(....)..).)))))).((((((((((((((.....))))))))))))))...... ( -35.30) >DroSec_CAF1 10541 116 - 1 GGCGGUC----GCAUCUCUGACAUCUGGAGAAGGAGCCAAGGCUGCCGGUAAUGGAGACUAGCCCGAAAGUACAUGCACCAGUAAGAGUAUUUUUCAGACGAAAAUAUUUUUGCAUAAAU (((((((----...(((((.......))))).(....)..)))))))(((.(((...(((........))).)))..))).((((((((((((((.....))))))))))))))...... ( -34.50) >DroSim_CAF1 12710 116 - 1 GGCGGUC----GCAUCUCUGACAUCUGGAGAAGGAGCCAAGGCUGCCGGUAAUGGAGACCAGCCCGAAAGUACAUGCACCAGUAAGAGUAUUUUUCAGACGAAAAUAUUUUUGCAUAAAU (((((((----...(((((.......))))).(((((....))).)).........)))).)))(....)...........((((((((((((((.....))))))))))))))...... ( -33.90) >DroEre_CAF1 2230 115 - 1 GGCGGCC----UCAUCUCCGCCAUCUGGAGAAGCAGCCAAGGCUGCCGGUAAUGGAGACCA-CCGGAAAGUACAUGCACCAGUGAGAGUAUUUUUCAGACGAAGAUAUUUUUGCAUAAAU (((((..----......))))).(((((....(((((....))))).(((.......))).-)))))..............(..(((((((((((.....)))))))))))..)...... ( -38.00) >DroYak_CAF1 11971 119 - 1 GGCGGUCGCUCGCAUCUCCGACAUCUGGAGAAGGAGCCAAGGCUGCCGGUAAUGGAGACCGGCCCGAAAGUACAUGCACCAGUAAGAGUAUUUUUCAGACGAAAAUAUU-UUGCAUAAAU (((((((((((.(.((((((.....)))))).)))))...)))))))(((...(....)..)))(....)...(((((.......((((((((((.....)))))))))-)))))).... ( -38.61) >consensus GGCGGUC____GCAUCUCCGACAUCUGGAGAAGGAGCCAAGGCUGCCGGUAAUGGAGACCAGCCCGAAAGUACAUGCACCAGUAAGAGUAUUUUUCAGACGAAAAUAUUUUUGCAUAAAU (((((((....((.((((((.....))))))....))...)))))))(((.(((...((..........)).)))..))).((((((((((((((.....))))))))))))))...... (-32.90 = -32.58 + -0.32)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:57:24 2006