
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
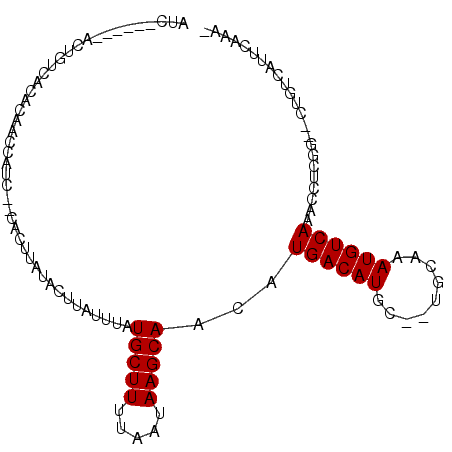
| Location | 26,058,399 – 26,058,522 |
| Length | 123 |
| Max. P | 0.726244 |

| Location | 26,058,399 – 26,058,492 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 82.49 |
| Mean single sequence MFE | -15.01 |
| Consensus MFE | -9.80 |
| Energy contribution | -9.80 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.68 |
| Structure conservation index | 0.65 |
| SVM decision value | -0.01 |
| SVM RNA-class probability | 0.529471 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 26058399 93 + 27905053 AUC------GCUGUCAUUCAACUAUC--CACUUAUACUUAUUUAUGCUUUUAAUAAGCAGCAUGACAUGC--UGCAAAUGUCAAACUUCGG--CUGUCAUUCAAA- ...------((((.............--................(((((.....)))))...((((((..--.....)))))).....)))--)...........- ( -14.80) >DroPse_CAF1 56038 95 + 1 CUC------AGAAUCCCACUAUCAUC--CACUUAUACUUAUUUAUGCUUUUAAUAAGCAGCAUGACAUGCUCUGCAAAUGUCAAACCUCGG--CUGUCAUUCAAA- ...------.((((..((((......--................(((((.....)))))...((((((.........))))))......))--.))..))))...- ( -13.70) >DroEre_CAF1 55492 93 + 1 AUC------GCUGCCAUUCAACCAUC--CACUUAUACUUAUUUAUGCUUUUAAUAAGCAACAUGACAUGC--UGCAAAUGUCAAACCUCGG--CUGUCAUUCAAA- ...------((.(((...........--................(((((.....)))))...((((((..--.....))))))......))--).))........- ( -16.40) >DroWil_CAF1 64837 93 + 1 ACC------CC---UACUAAACUAUC--CACUUAUGCUUAUUUAUGCUUUUAAUAAGCAACAUGACAUGC--UGCAAAUGUCAAUACUCGGUCCUGUCAUUCAAAA (((------..---............--......((((((((.........))))))))...((((((..--.....))))))......))).............. ( -14.40) >DroAna_CAF1 54012 101 + 1 GUCUCCCCCUCUGCGAUAUAAGUACUGACACUUAUACUUAUUUAUGCUUUUAAUAAGCAACAUGACAUGC--UGCAAAUGUCAAACCUCGG--CUGUCAUUCAAG- ................(((((((......)))))))........(((((.....)))))..((((((.((--((..............)))--))))))).....- ( -17.04) >DroPer_CAF1 58644 95 + 1 CUC------AGAAUCCCACUAUCAUC--CACUUAUACUUAUUUAUGCUUUUAAUAAGCAGCAUGACAUGCUCUGCAAAUGUCAAACCUCGG--CUGUCAUUCAAA- ...------.((((..((((......--................(((((.....)))))...((((((.........))))))......))--.))..))))...- ( -13.70) >consensus AUC______ACUGUCACACAACCAUC__CACUUAUACUUAUUUAUGCUUUUAAUAAGCAACAUGACAUGC__UGCAAAUGUCAAACCUCGG__CUGUCAUUCAAA_ ............................................(((((.....)))))...((((((.........))))))....................... ( -9.80 = -9.80 + -0.00)


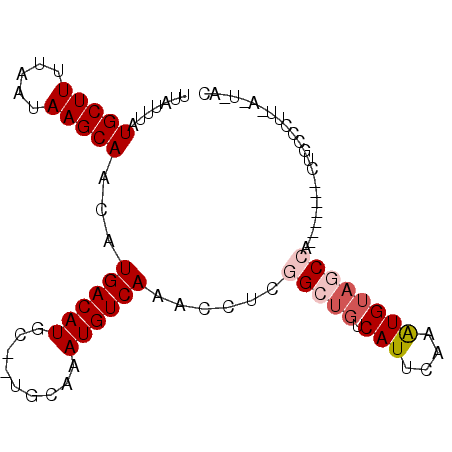
| Location | 26,058,428 – 26,058,522 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 96 |
| Reading direction | forward |
| Mean pairwise identity | 83.82 |
| Mean single sequence MFE | -17.15 |
| Consensus MFE | -12.92 |
| Energy contribution | -13.78 |
| Covariance contribution | 0.86 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.18 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.41 |
| SVM RNA-class probability | 0.726244 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 26058428 94 + 27905053 UUAUUUAUGCUUUUAAUAAGCAGCAUGACAUGC--UGCAAAUGUCAAACUUCGGCUGUCAUUCAAAUGUAGCCAUGCAGCACUGCCCUUCACUUCC .......(((((.....)))))(((.(...(((--((((...(........)(((((.(((....)))))))).)))))))))))........... ( -22.60) >DroPse_CAF1 56067 91 + 1 UUAUUUAUGCUUUUAAUAAGCAGCAUGACAUGCUCUGCAAAUGUCAAACCUCGGCUGUCAUUCAAAUGUUACAU-----UUCUCCCCCUUAUUCAC .(((((.(((((.....)))))(.(((((((((...)))...(((.......))))))))).))))))......-----................. ( -11.90) >DroEre_CAF1 55521 80 + 1 UUAUUUAUGCUUUUAAUAAGCAACAUGACAUGC--UGCAAAUGUCAAACCUCGGCUGUCAUUCAAAUGUAGCCA-------CUGCCU-------GC .......(((((.....)))))...((((((..--.....))))))......(((((.(((....)))))))).-------......-------.. ( -18.70) >DroYak_CAF1 52867 87 + 1 UUAUUUAUGCUUUUAAUAAGCAACAUGACAUGC--UGCAAAUGUCAAACCUCGGCUGUCAUUCAAAUGUAGCCA-------CUGCCCUUCACUUUC .......(((((.....)))))...((((((..--.....))))))......(((((.(((....)))))))).-------............... ( -18.70) >DroAna_CAF1 54049 81 + 1 UUAUUUAUGCUUUUAAUAAGCAACAUGACAUGC--UGCAAAUGUCAAACCUCGGCUGUCAUUCAAGUGAAGCCA-------CUACUCU------CC .......(((((.....)))))...((((((..--.....))))))......((((.((((....)))))))).-------.......------.. ( -19.10) >DroPer_CAF1 58673 91 + 1 UUAUUUAUGCUUUUAAUAAGCAGCAUGACAUGCUCUGCAAAUGUCAAACCUCGGCUGUCAUUCAAAUGUAACAU-----UUCUCCCCCUUAUUCAC .(((((.(((((.....)))))(.(((((((((...)))...(((.......))))))))).))))))......-----................. ( -11.90) >consensus UUAUUUAUGCUUUUAAUAAGCAACAUGACAUGC__UGCAAAUGUCAAACCUCGGCUGUCAUUCAAAUGUAGCCA_______CUGCCCUU_A_U_AC .......(((((.....)))))...((((((.........))))))......(((((.(((....))))))))....................... (-12.92 = -13.78 + 0.86)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:42:35 2006