
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 26,048,990 – 26,049,128 |
| Length | 138 |
| Max. P | 0.685633 |

| Location | 26,048,990 – 26,049,092 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 79.94 |
| Mean single sequence MFE | -40.55 |
| Consensus MFE | -19.90 |
| Energy contribution | -19.10 |
| Covariance contribution | -0.80 |
| Combinations/Pair | 1.26 |
| Mean z-score | -2.11 |
| Structure conservation index | 0.49 |
| SVM decision value | -0.04 |
| SVM RNA-class probability | 0.513176 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 26048990 102 - 27905053 CAUCGCACUGGUGGCAACCCUGGUCAGCGG--GGAGGCCCGGCAAUCCAGGCAGCUGGUCAACCUGUUUAAGGUUGCAUACUCAUGCGACAAAUGUGCCAAGCA ....((..(((((((...(((((...((.(--((...))).))...)))))..))).((((((((.....)))))((((....)))))))......)))).)). ( -36.40) >DroPse_CAF1 46236 101 - 1 CGUGGAGCUGGUGGCAGCUCCGGUCGGCGCCAGAAUG---GAGAAUGCCGCCAGCUGGGCAACCUGUUUGAUGUUGCAUGUUCAUGUGACAAAUGUUGCACGCA .(((.(((((((((((.(((((.((.......)).))---)))..)))))))))))..(((((..(((...(((..(((....)))..))))))))))).))). ( -45.20) >DroSim_CAF1 42907 102 - 1 CAUCGCACUGGUGGCAACCCUGGUCAGCGG--GGAAGCCCGGCAAUCCAGGCAGCUGGUCAACCUGUUUGAGGUUGCAUACUCAUGCGACAAAUGUGCCACGCA ....((....((((((......(((..(((--(....))))(((((((((((((.(.....).))))))).))))))..........))).....)))))))). ( -38.20) >DroEre_CAF1 46190 101 - 1 CGUCGCACUGGUGGCAACCCUGGCCAGCGG--GCAUGC-AGGCUAUCCAGGCAGCUGGUCAACCUGUUUGAGGUUGCAUACUCAUGCGACAAAUGUGCCACGCA .((((((..((((((((((...(((....)--))..((-(((....((((....))))....)))))....)))))).))))..))))))...((((...)))) ( -40.00) >DroYak_CAF1 43031 97 - 1 CGUCGCACUGGUGGCAACCCUGGCAACC------CUGG-CAGCUAUUCGGGCAGCUGGCCAACCUGUUUGAGGUUGCAUACUCAUGCGACAAAUGUGCCACGCA .((((((..(((((((((((..(((...------.(((-(((((........)))).))))...)))..).)))))).))))..))))))...((((...)))) ( -37.10) >DroPer_CAF1 48886 101 - 1 CGUGGAGCUGGUGGCAGCUCCGGUCGGCGCCAGAAUG---GAGAAUGCCGCCAGCAGGGCAACCUGUUUGAUGUUGCAUGUUCAUGUGACAAAUGUUGCACGCA ((((.(((((((((((.(((((.((.......)).))---)))..))))))))(((((....)))))....(((..(((....)))..)))...))).)))).. ( -46.40) >consensus CGUCGCACUGGUGGCAACCCUGGUCAGCGG__GAAUGC_CGGCAAUCCAGGCAGCUGGUCAACCUGUUUGAGGUUGCAUACUCAUGCGACAAAUGUGCCACGCA (((.((((.((((((.......)))........................(((.....))).)))........(((((((....)))))))....)))).))).. (-19.90 = -19.10 + -0.80)

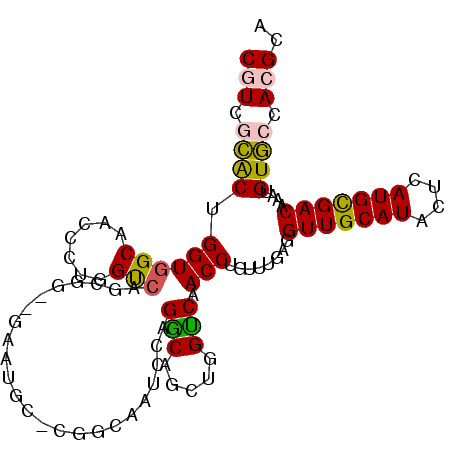

| Location | 26,049,014 – 26,049,128 |
|---|---|
| Length | 114 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 81.15 |
| Mean single sequence MFE | -44.98 |
| Consensus MFE | -20.94 |
| Energy contribution | -20.33 |
| Covariance contribution | -0.61 |
| Combinations/Pair | 1.20 |
| Mean z-score | -2.76 |
| Structure conservation index | 0.47 |
| SVM decision value | 0.32 |
| SVM RNA-class probability | 0.685633 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 26049014 114 + 27905053 UAUGCAACCUUAAACAGGUUGACCAGCUGCCUGGAUUGCCGGGCCUCC--CCGCUGACCAGGGUUGCCACCAGUGCGAUGGCAACUGUCAAAUUUGACAAAUGACUG---GUCAAGCGG ....((((((.....)))))).......((((((....))))))....--(((((((((((((((((((.(.....).))))))))((((....))))......)))---))).))))) ( -50.10) >DroPse_CAF1 46260 113 + 1 CAUGCAACAUCAAACAGGUUGCCCAGCUGGCGGCAUUCUC---CAUUCUGGCGCCGACCGGAGCUGCCACCAGCUCCACGAGAGCUGUCAGAUUUGACAAACGACUG---GUCAAGCGG ...(((((.........)))))((.(((((((.((.....---.....)).))))(((((((((((....)))))))........((((......)))).......)---))).))))) ( -39.80) >DroSim_CAF1 42931 114 + 1 UAUGCAACCUCAAACAGGUUGACCAGCUGCCUGGAUUGCCGGGCUUCC--CCGCUGACCAGGGUUGCCACCAGUGCGAUGGCAACUGUCAAAUUUGACAAAUGACUG---GUCAAGCGG ....((((((.....)))))).......((((((....))))))....--(((((((((((((((((((.(.....).))))))))((((....))))......)))---))).))))) ( -50.50) >DroEre_CAF1 46214 113 + 1 UAUGCAACCUCAAACAGGUUGACCAGCUGCCUGGAUAGCCU-GCAUGC--CCGCUGGCCAGGGUUGCCACCAGUGCGACGGCAACUGUCAAAUUUGACAAAUGACUG---GUCAAGCGG ..............(((((((.((((....)))).))))))-).....--((((((((((((((((((...........)))))))((((....))))......)))---))).))))) ( -45.10) >DroYak_CAF1 43055 112 + 1 UAUGCAACCUCAAACAGGUUGGCCAGCUGCCCGAAUAGCUG-CCAG------GGUUGCCAGGGUUGCCACCAGUGCGACGGCAACUGUCAAAUUUGACAAAUGACUGGUGGUCAAGCGG ...(((((((.........((((.(((((......))))))-))))------))))))...(.(((((((((((((....))...(((((....)))))....)))))))).))).).. ( -43.00) >DroPer_CAF1 48910 113 + 1 CAUGCAACAUCAAACAGGUUGCCCUGCUGGCGGCAUUCUC---CAUUCUGGCGCCGACCGGAGCUGCCACCAGCUCCACGAGAGCUGUCAGAUUUGACAAACGACUG---GUCAAGCGG ...(((((.........))))).(((((((((.((.....---.....)).))))(((((((((((....)))))))........((((......)))).......)---))).))))) ( -41.40) >consensus UAUGCAACCUCAAACAGGUUGACCAGCUGCCUGGAUUGCCG_GCAUCC__CCGCUGACCAGGGUUGCCACCAGUGCGACGGCAACUGUCAAAUUUGACAAAUGACUG___GUCAAGCGG ....((((((.....)))))).((.(((((.......))................(((((((((((((...........)))))))((((....))))......)))...))).))))) (-20.94 = -20.33 + -0.61)

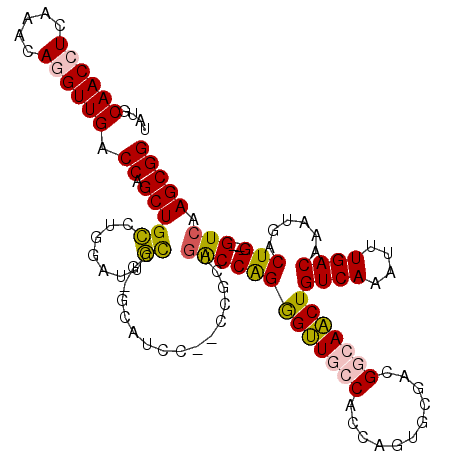

| Location | 26,049,014 – 26,049,128 |
|---|---|
| Length | 114 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 81.15 |
| Mean single sequence MFE | -46.10 |
| Consensus MFE | -22.97 |
| Energy contribution | -23.87 |
| Covariance contribution | 0.89 |
| Combinations/Pair | 1.22 |
| Mean z-score | -2.41 |
| Structure conservation index | 0.50 |
| SVM decision value | 0.11 |
| SVM RNA-class probability | 0.587570 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 26049014 114 - 27905053 CCGCUUGAC---CAGUCAUUUGUCAAAUUUGACAGUUGCCAUCGCACUGGUGGCAACCCUGGUCAGCGG--GGAGGCCCGGCAAUCCAGGCAGCUGGUCAACCUGUUUAAGGUUGCAUA (((((.(((---(((.....(((((....)))))(((((((((.....))))))))).)))))))))))--((..(((.((....)).)))..)).(.((((((.....)))))))... ( -52.20) >DroPse_CAF1 46260 113 - 1 CCGCUUGAC---CAGUCGUUUGUCAAAUCUGACAGCUCUCGUGGAGCUGGUGGCAGCUCCGGUCGGCGCCAGAAUG---GAGAAUGCCGCCAGCUGGGCAACCUGUUUGAUGUUGCAUG ((((..((.---...))..((((((....)))))).....))))(((((((((((.(((((.((.......)).))---)))..)))))))))))..(((((.........)))))... ( -44.40) >DroSim_CAF1 42931 114 - 1 CCGCUUGAC---CAGUCAUUUGUCAAAUUUGACAGUUGCCAUCGCACUGGUGGCAACCCUGGUCAGCGG--GGAAGCCCGGCAAUCCAGGCAGCUGGUCAACCUGUUUGAGGUUGCAUA (((((.(((---(((.....(((((....)))))(((((((((.....))))))))).)))))))))))--((....)).(((((((((((((.(.....).))))))).))))))... ( -52.60) >DroEre_CAF1 46214 113 - 1 CCGCUUGAC---CAGUCAUUUGUCAAAUUUGACAGUUGCCGUCGCACUGGUGGCAACCCUGGCCAGCGG--GCAUGC-AGGCUAUCCAGGCAGCUGGUCAACCUGUUUGAGGUUGCAUA ..((..(((---((((....(((((....)))))((((((..(.....)..))))))(((((..(((..--(....)-..)))..)))))..)))))))(((((.....)))))))... ( -44.00) >DroYak_CAF1 43055 112 - 1 CCGCUUGACCACCAGUCAUUUGUCAAAUUUGACAGUUGCCGUCGCACUGGUGGCAACCCUGGCAACC------CUGG-CAGCUAUUCGGGCAGCUGGCCAACCUGUUUGAGGUUGCAUA ....(((.((((((((...((((((....))))))..((....))))))))))))).....((((((------(..(-(((.......(((.....)))...))))..).))))))... ( -38.80) >DroPer_CAF1 48910 113 - 1 CCGCUUGAC---CAGUCGUUUGUCAAAUCUGACAGCUCUCGUGGAGCUGGUGGCAGCUCCGGUCGGCGCCAGAAUG---GAGAAUGCCGCCAGCAGGGCAACCUGUUUGAUGUUGCAUG ..((((.((---.((..(((.((((....))))))).)).)).))))((((((((.(((((.((.......)).))---)))..))))))))(((((....)))))............. ( -44.60) >consensus CCGCUUGAC___CAGUCAUUUGUCAAAUUUGACAGUUGCCGUCGCACUGGUGGCAACCCUGGUCAGCGG__GAAUGC_CGGCAAUCCAGGCAGCUGGUCAACCUGUUUGAGGUUGCAUA .((((((((.....))))..(((((....)))))(((((((((.....))))))))).......))))............((((((((((((...((....)))))))).))))))... (-22.97 = -23.87 + 0.89)


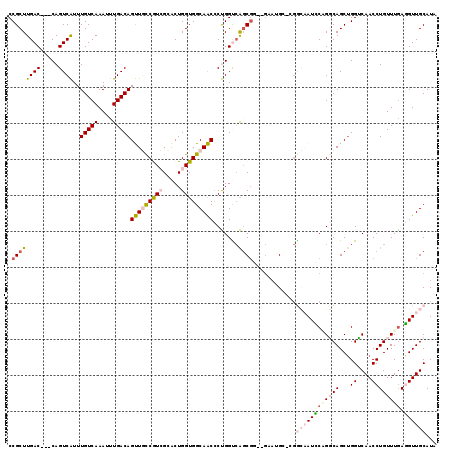
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:42:34 2006