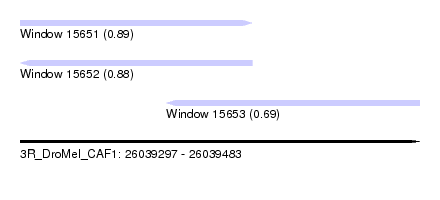
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 26,039,297 – 26,039,483 |
| Length | 186 |
| Max. P | 0.893586 |

| Location | 26,039,297 – 26,039,405 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 85.79 |
| Mean single sequence MFE | -24.63 |
| Consensus MFE | -20.26 |
| Energy contribution | -19.86 |
| Covariance contribution | -0.40 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.92 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.98 |
| SVM RNA-class probability | 0.893586 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 26039297 108 + 27905053 GAAAA----UCGA-AAUUAAUAAUUCAUUUUGUUUAGUUUUGAAGGGAAUCCAUACGAUUUUUUUUCCCAGCUUCCUAAGCCAACCAGCCUCUGAGUUUCCCAGGCUGAUGGC .....----((((-(((((((((......))).)))))))))).(((((...............)))))..........((((..((((((..(.....)..)))))).)))) ( -26.26) >DroSec_CAF1 32158 109 + 1 GAAAA----UCCAAAAUAAAUAAUUCAUUUUGUUUAGCUUAGAAGGGAAUCCAUACGAUUUUCUUUCCCAGCUUCCUAAGCCAACCAGCCUCUGUGUUUCCCAGGCUGAUGGC .....----.......(((((((......)))))))(((..((((..((((.....))))..))))...))).......((((..((((((..(.....)..)))))).)))) ( -26.20) >DroSim_CAF1 33543 108 + 1 GAAAA----UCGA-GUUUAAUAAUUCAUUUCAUUUAGUUUAGAAGGGAAUCCAUACGAUUUUCUUUCCCAGCUUCCUAAGCCAACCAGCCUCUGUGUUUCCCAGGCUGAUGGC ((((.----..((-(((....))))).))))....((((..((((..((((.....))))..))))...))))......((((..((((((..(.....)..)))))).)))) ( -24.60) >DroEre_CAF1 37358 110 + 1 GAAAAUACGUCCA-AAUUAGUAUUGUAUUUC--UUAAUUUAGAAAGAAAUUCUUACGAUUUUCUUGCCCAGCUUCCUAAGCCAACCAGCCUCCGAAUUUCCCAGGCUGAUGGC ((((((.(((...-....((.(((...((((--(......)))))..))).)).)))))))))................((((..((((((..(.....)..)))))).)))) ( -21.10) >DroYak_CAF1 32925 106 + 1 GAAAA----CCCA-AAUAAAUAUUUUAUUUC--GUAGCUUAGAAAGGAAUCCUUACGAUUUUCUUUCCCAGCUUCCUGAGCCAACCAGCCUCCGAACUUCCCAGGCUGAUGGC .....----....-...............((--(.((((..((((((((..(....)..))))))))..))))...)))((((..((((((..(.....)..)))))).)))) ( -25.00) >consensus GAAAA____UCCA_AAUUAAUAAUUCAUUUC_UUUAGUUUAGAAGGGAAUCCAUACGAUUUUCUUUCCCAGCUUCCUAAGCCAACCAGCCUCUGAGUUUCCCAGGCUGAUGGC ...................................((((..((((((((..(....)..))))))))..))))......((((..((((((..(.....)..)))))).)))) (-20.26 = -19.86 + -0.40)

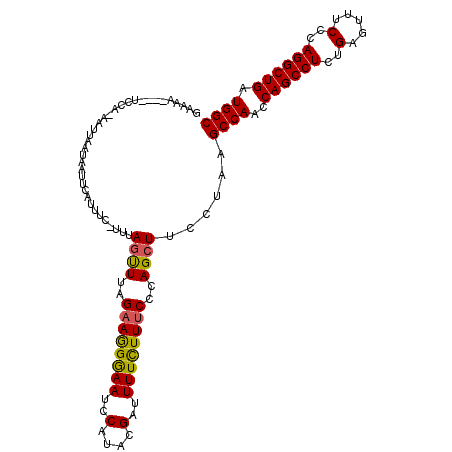
| Location | 26,039,297 – 26,039,405 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 85.79 |
| Mean single sequence MFE | -31.16 |
| Consensus MFE | -21.20 |
| Energy contribution | -21.44 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.47 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.89 |
| SVM RNA-class probability | 0.875117 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 26039297 108 - 27905053 GCCAUCAGCCUGGGAAACUCAGAGGCUGGUUGGCUUAGGAAGCUGGGAAAAAAAAUCGUAUGGAUUCCCUUCAAAACUAAACAAAAUGAAUUAUUAAUU-UCGA----UUUUC (((((((((((((....))...))))))).))))((((......(((((...............))))).......))))......((((........)-))).----..... ( -27.08) >DroSec_CAF1 32158 109 - 1 GCCAUCAGCCUGGGAAACACAGAGGCUGGUUGGCUUAGGAAGCUGGGAAAGAAAAUCGUAUGGAUUCCCUUCUAAGCUAAACAAAAUGAAUUAUUUAUUUUGGA----UUUUC (((((((((((.(....)....))))))).))))((((....))))....(((((((((.((((......)))).))....((((((((.....))))))))))----))))) ( -34.40) >DroSim_CAF1 33543 108 - 1 GCCAUCAGCCUGGGAAACACAGAGGCUGGUUGGCUUAGGAAGCUGGGAAAGAAAAUCGUAUGGAUUCCCUUCUAAACUAAAUGAAAUGAAUUAUUAAAC-UCGA----UUUUC (((((((((((.(....)....))))))).))))((((....))))....((((((((....(((((..(((..........)))..))))).......-.)))----))))) ( -30.50) >DroEre_CAF1 37358 110 - 1 GCCAUCAGCCUGGGAAAUUCGGAGGCUGGUUGGCUUAGGAAGCUGGGCAAGAAAAUCGUAAGAAUUUCUUUCUAAAUUAA--GAAAUACAAUACUAAUU-UGGACGUAUUUUC .(.((((((((..(.....)..)))))))).)((((((....))))))((((((.((....)).))))))(((((((((.--............)))))-))))......... ( -30.72) >DroYak_CAF1 32925 106 - 1 GCCAUCAGCCUGGGAAGUUCGGAGGCUGGUUGGCUCAGGAAGCUGGGAAAGAAAAUCGUAAGGAUUCCUUUCUAAGCUAC--GAAAUAAAAUAUUUAUU-UGGG----UUUUC (((((((((((..(.....)..))))))).))))......((((.((((((.((.((....)).)).)))))).)))).(--.((((((.....)))))-).).----..... ( -33.10) >consensus GCCAUCAGCCUGGGAAACUCAGAGGCUGGUUGGCUUAGGAAGCUGGGAAAGAAAAUCGUAUGGAUUCCCUUCUAAACUAAA_GAAAUGAAUUAUUAAUU_UGGA____UUUUC (((((((((((..(.....)..))))))).))))((((..((..(((((...............)))))..))...))))................................. (-21.20 = -21.44 + 0.24)

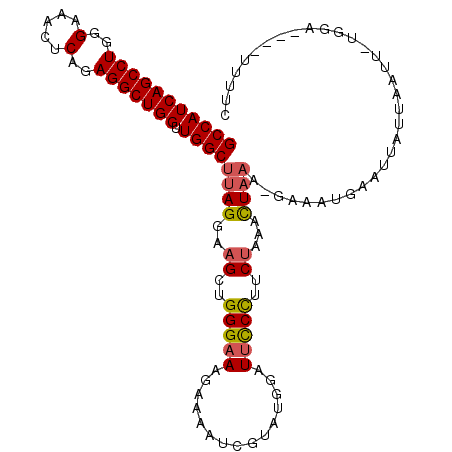
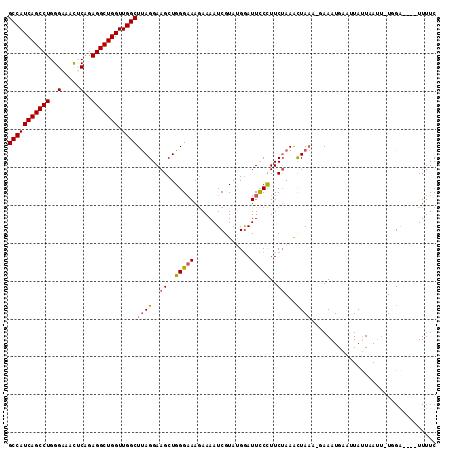
| Location | 26,039,365 – 26,039,483 |
|---|---|
| Length | 118 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 84.10 |
| Mean single sequence MFE | -34.67 |
| Consensus MFE | -27.21 |
| Energy contribution | -27.93 |
| Covariance contribution | 0.73 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.92 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.32 |
| SVM RNA-class probability | 0.685113 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 26039365 118 - 27905053 UUUCUU--UCAAUCCGAUCGUAAGCCAACAAUCUCGUCAUGAACUUCAACAAGAUCUUCGUCUUUGUGGCACUCAUCCUGGCCAUCAGCCUGGGAAACUCAGAGGCUGGUUGGCUUAGGA ......--.........((.(((((((((.........(((((..((.....))..)))))....(((((.(.......))))))((((((((....))...))))))))))))))).)) ( -36.70) >DroSec_CAF1 32227 118 - 1 UUUCUU--GCAAUCCGGUCGUAAGCCAACAAUCUCGUCAUGAACUUCAACAAGAUCUUCGUCUUCGUGGCCCUCAUCCUGGCCAUCAGCCUGGGAAACACAGAGGCUGGUUGGCUUAGGA ......--.........((.(((((((((.........(((((..((.....))..)))))....((((((........))))))((((((.(....)....))))))))))))))).)) ( -38.80) >DroSim_CAF1 33611 118 - 1 UUUCUU--UCAAUCCAGUCGUAAGCCAACAAUCUCGUCAUGAACUUCAACAAGAUCUUCGUCUUCGUGGCCCUCAUCCUGGCCAUCAGCCUGGGAAACACAGAGGCUGGUUGGCUUAGGA ......--.........((.(((((((((.........(((((..((.....))..)))))....((((((........))))))((((((.(....)....))))))))))))))).)) ( -38.80) >DroEre_CAF1 37428 118 - 1 UUCCUA--UCAAUCCGGUAACAAGCCGACAAUCCCGUCAUGAACUUCAACAAGAUCUUCGUCUUCGUGGCCCUCAUCCUGGCCAUCAGCCUGGGAAAUUCGGAGGCUGGUUGGCUUAGGA .(((((--((((.(((((......((((...(((((...(((......(((((((....))))).))((((........)))).)))...)))))...))))..)))))))))..))))) ( -36.90) >DroYak_CAF1 32991 118 - 1 CUCCUC--UCAAUUCAAUAAUAAGCCAACAAUCUAGACAUGAACUUCAACAAGAUCUUCGUCUUCGUGGCUCUCAUCUUGGCCAUCAGCCUGGGAAGUUCGGAGGCUGGUUGGCUCAGGA ......--..............(((((.......((((..(((..((.....))..)))))))...)))))....((((((((((((((((..(.....)..))))))).)))).))))) ( -33.60) >DroAna_CAF1 33989 115 - 1 UUACUUUCUGAAUCAA-----AGAAAAAUAUUCCCAUCAUGAACUUCAACAAGAUCUUCGUCUUCGUCGCCAUUAUCCUGGCCAUUAGCUUGGGACAGACCGAAGCAGGUUGGCUGAAAA ....(((((.......-----))))).....(((((..(((((..((.....))..))))).......((((......))))........)))))(((.((((......))))))).... ( -23.20) >consensus UUUCUU__UCAAUCCAGUCGUAAGCCAACAAUCUCGUCAUGAACUUCAACAAGAUCUUCGUCUUCGUGGCCCUCAUCCUGGCCAUCAGCCUGGGAAACACAGAGGCUGGUUGGCUUAGGA ....................(((((((((.........(((((..((.....))..)))))....((((((........))))))((((((..(.....)..)))))))))))))))... (-27.21 = -27.93 + 0.73)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:42:29 2006