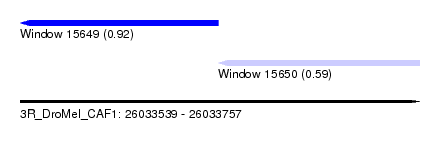
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 26,033,539 – 26,033,757 |
| Length | 218 |
| Max. P | 0.922356 |

| Location | 26,033,539 – 26,033,647 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 79.22 |
| Mean single sequence MFE | -25.20 |
| Consensus MFE | -20.15 |
| Energy contribution | -19.60 |
| Covariance contribution | -0.55 |
| Combinations/Pair | 1.30 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.80 |
| SVM decision value | 1.15 |
| SVM RNA-class probability | 0.922356 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
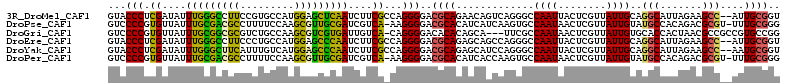
>3R_DroMel_CAF1 26033539 108 - 27905053 CCUGCAUCUCCAAAUAUUGGCCACAGU-UGGUCUUUUGAUAC-----AUCGUUCUUGUGUUUUUCGCUGGCUG-UCCUUCCAAAGCCAGUGAUUGUCAAACAGAACGUACAUUAA ..........((((....(((((....-))))).))))....-----..((((((..((....(((((((((.-.........)))))))))....))...))))))........ ( -29.50) >DroPse_CAF1 30036 106 - 1 CUUGCAUCUCCAUAUAUUGGCCACAGU-----C---UGGUACCCUUGACCGUUCUUGUGUUAUUCGCUAGACU-UACCUUCUAGUCCAGCGAUUGUCAAACAGAACGUACACUAA ...................((((....-----.---)))).....((..((((((..((.((.(((((.((((-........)))).))))).)).))...))))))..)).... ( -23.90) >DroEre_CAF1 27749 105 - 1 CCUGCAUCUCCAAAUAUUGGUCACAGU-UGAUC---UGAUAC-----ACCGUUCUUGUGUUUUUCGCUGGUUG-UCUCACCAAAACCAGUGAUUGUCAAACAGAACGUAAAUUAA ..............(((..((((....-))).)---..))).-----..((((((..((....(((((((((.-.........)))))))))....))...))))))........ ( -24.30) >DroYak_CAF1 27213 105 - 1 CCUGCAUCUCCAAAUAUUGGCCACACU-UGGUC---UGAUAC-----ACCGUUCUUGUGUUUUUCGCUGGUUG-UCCUACCAAAACCAGGGAUUGUCAAACAGAACGCAAAUUAA ..(((.........(((..((((....-))).)---..))).-----...(((((..((....((.((((((.-.........)))))).))....))...))))))))...... ( -26.20) >DroAna_CAF1 29191 107 - 1 CCUGCAUCUCCAUAUAUUGGCCACAUUCUGGUC---UAUCCG-----UCCGUUCUUGUGUUAUUUGCUGGAUAAUCUCCUCAAGACCAGUGAUUGUCAAAAAGAACGAAGACUAA ..................(((((.....)))))---.....(-----(((((((((.((.((.(..((((....(((.....)))))))..).)).))..)))))))..)))... ( -23.40) >DroPer_CAF1 30684 106 - 1 CUUGCAUCUCCAUAUAUUGGCCACAGU-----C---UGGUACCCUUGACCGUUCUUGUGUUAUUCGCUAGACU-UACCUUCUAGUCCAGCGAUUGUCAAACAGAACGUACACUAA ...................((((....-----.---)))).....((..((((((..((.((.(((((.((((-........)))).))))).)).))...))))))..)).... ( -23.90) >consensus CCUGCAUCUCCAAAUAUUGGCCACAGU_UGGUC___UGAUAC_____ACCGUUCUUGUGUUAUUCGCUGGAUG_UCCCUCCAAAACCAGUGAUUGUCAAACAGAACGUACACUAA .........(((.....))).............................((((((..((....(((((((((...........)))))))))....))...))))))........ (-20.15 = -19.60 + -0.55)

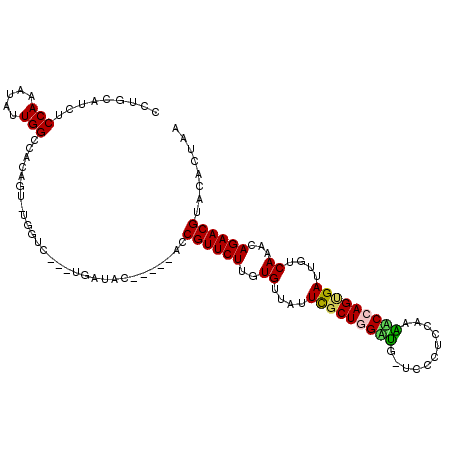
| Location | 26,033,647 – 26,033,757 |
|---|---|
| Length | 110 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 72.96 |
| Mean single sequence MFE | -37.02 |
| Consensus MFE | -23.16 |
| Energy contribution | -20.72 |
| Covariance contribution | -2.44 |
| Combinations/Pair | 1.48 |
| Mean z-score | -1.19 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.593514 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 26033647 110 - 27905053 GUACCCUCGAUAUUUGGGCCUUCCGUGCCAUGGAGCUCAAUCUUCGCCAGGGGACGCAGAACAGUCAGGGCCAAUUACUCGUUAUUGCAGGCAUUAGAAGCC--AUUGCGGU ...(((((((...((((((..(((((...)))))))))))...)))..))))..(((((..((((..(((.......)))...))))..(((.......)))--.))))).. ( -31.40) >DroPse_CAF1 30142 110 - 1 GUCCCCGUGUUAUUUGCGACGCCUUUUCCAAGCGUUGCGAUCGUCA-AAGGGGACGCACAUCAUCAAGUGCCAAUAACUCGUUAUUGUAUGCCACAGACGCGU-UUUGCGGG ((((((.((....(((((((((.........)))))))))....))-..))))))((((........))))((((((....))))))....((.((((.....-)))).)). ( -37.20) >DroGri_CAF1 37282 108 - 1 GUCCCCGUGUUAUUUGCGGCGCGUCUGCCAAGCGUCGUGAUUGUCA-CAGGGGACACACAGCA---UUCGCCAAUAACUCGUUAUUGUGCACCACUAACGCCGCCGUGCCGG (((((((((.((.(..((((((.........))))))..).)).))-).)))))).(((.((.---...((((((((....)))))).))....(....)..)).))).... ( -41.10) >DroEre_CAF1 27854 110 - 1 GUACCCUCGAUAUUUGGGCCUUCCCUGCCAUGGAGCCCAAUCUUCGCCAGGGGACGCAGAGCAGCCAGGGCCAAUUACUCGUUAUUGCAGGCAUUAGAAGCC--AUUGCGGU ...(((((((...((((((..(((.......)))))))))...)))..))))..(((((.((((...(((.......)))....)))).(((.......)))--.))))).. ( -35.30) >DroYak_CAF1 27318 110 - 1 GUACCCUCGAUAUUUGGGCUUCAUUUGUCAUGGAGCCCAAUCUUCGCCAGGGGACGCAGAGCAUCCAGGGCCAAUUACUCGUUAUUGCAGGCAUUAGAAGCC--AAUGCGGU ...(((((((...(((((((((((.....)))))))))))...)))..))))..((((..(((....(((.......))).....))).(((.......)))--..)))).. ( -39.90) >DroPer_CAF1 30790 110 - 1 GUCCCCGUGUUAUUUGCGACGCCUUUUCCAAGCGUUGCGAUCGUCA-AAGGGGACGCACAUCACCAAGUGCCAAUAACUCGUUAUUGUAUGCCACAGACGCGU-UUUGCGGG ((((((.((....(((((((((.........)))))))))....))-..))))))((((........))))((((((....))))))....((.((((.....-)))).)). ( -37.20) >consensus GUACCCGCGAUAUUUGCGCCGCCUUUGCCAAGCAGCGCAAUCGUCA_CAGGGGACGCACAGCAGCCAGGGCCAAUAACUCGUUAUUGCAGGCAACAGAAGCC__AUUGCGGG ...(((.((....(((((((((.........)))))))))....))...)))..((((.............((((........))))..(((.......)))....)))).. (-23.16 = -20.72 + -2.44)
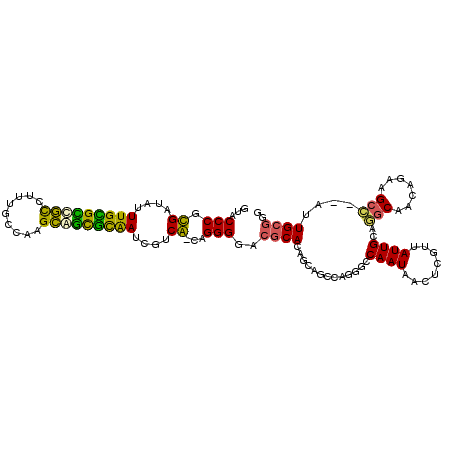
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:42:27 2006