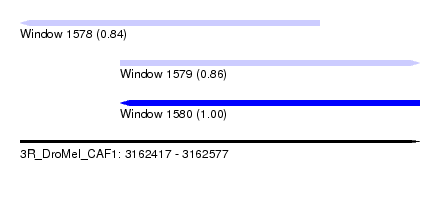
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 3,162,417 – 3,162,577 |
| Length | 160 |
| Max. P | 0.999034 |

| Location | 3,162,417 – 3,162,537 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 76.18 |
| Mean single sequence MFE | -20.46 |
| Consensus MFE | -13.68 |
| Energy contribution | -14.42 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.73 |
| SVM RNA-class probability | 0.835223 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 3162417 120 - 27905053 AAGCGGAAAUAAAGAGGCUCACAGUGUAUUUUGUAGUGUACUGUGAAGUUCUUAAUGCAAUGAAUAAAAAAAAAACACCAAAAAACUUUGAAUAAAAAUUAACUAAAUUACAAUCAACUA .((..((....(((((..((((((((((((....))))))))))))..))))).........................((((....)))).......................))..)). ( -17.50) >DroSec_CAF1 10305 118 - 1 AAGCGGAAAUAAAGAGGCUUACAGUGUAUUUUGAAGUGCACUGUGAAGUUCUUAAGACAAUGAAUAGGAAAAA-AC-CCAAAAAACCUUUAAUAAUAAUUAGAUAUAUUAUAAUCAACCA ....((.....(((((..((((((((((((....))))))))))))..))))).......(((...((.....-..-))............((((((........))))))..))).)). ( -22.90) >DroSim_CAF1 12473 119 - 1 AAGCGGAAAUAAAGAGGCUCACAGUGUAUUUUGUAGUGCACCGUGAAGUUCUUAAGACAAUGAAUAGGAAAAAUAC-UCAAAAAACCUUUAAUAAAAAUUACCUAAAUUAUAAUUACCCA ....((.....(((((..((((.(((((((....))))))).))))..)))))...........((((........-........................))))............)). ( -18.65) >DroEre_CAF1 2013 107 - 1 AAGCGGAAAUAAAGAGGCUCACAGUGUAUUUUGUACUGCACUGUCAGAACUUUGAGCCAAGCCA------------ACU-AAAAUACUUUAUUAGAAAUAAACUAAGUUGGAUUAAAUCA .............(((((((((((((((........)))))))).........)))))...(((------------(((-....((.(((((.....))))).))))))))......)). ( -22.80) >consensus AAGCGGAAAUAAAGAGGCUCACAGUGUAUUUUGUAGUGCACUGUGAAGUUCUUAAGACAAUGAAUAGGAAAAA_AC_CCAAAAAACCUUUAAUAAAAAUUAACUAAAUUAUAAUCAACCA ...........(((((..((((((((((((....))))))))))))..)))))................................................................... (-13.68 = -14.42 + 0.75)

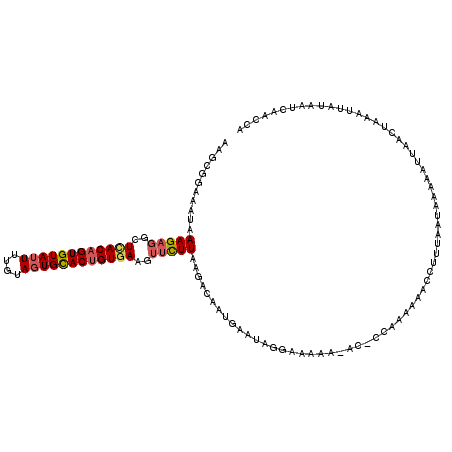
| Location | 3,162,457 – 3,162,577 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 77.83 |
| Mean single sequence MFE | -21.31 |
| Consensus MFE | -14.60 |
| Energy contribution | -15.44 |
| Covariance contribution | 0.84 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.78 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.83 |
| SVM RNA-class probability | 0.860797 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 3162457 120 + 27905053 UGGUGUUUUUUUUUUAUUCAUUGCAUUAAGAACUUCACAGUACACUACAAAAUACACUGUGAGCCUCUUUAUUUCCGCUUAGAUAAGUUUACUUCAUAAGCGACACUAAAUUAUAUGCCA ((((((.....................((((..((((((((..............))))))))..))))......((((((...(((....)))..))))))))))))............ ( -23.54) >DroSec_CAF1 10345 118 + 1 UGG-GU-UUUUUCCUAUUCAUUGUCUUAAGAACUUCACAGUGCACUUCAAAAUACACUGUAAGCCUCUUUAUUUCCGCUUAGAUAAGUUUACUUCAUAAGCGACACUAAAUUAUAUGCCA (((-(.-.....)))).....((((..((((.(((.((((((............)))))))))..)))).......(((((...(((....)))..)))))))))............... ( -21.10) >DroSim_CAF1 12513 119 + 1 UGA-GUAUUUUUCCUAUUCAUUGUCUUAAGAACUUCACGGUGCACUACAAAAUACACUGUGAGCCUCUUUAUUUCCGCUUAGAUAAGUUUACUUCAUAAGCGACACUAAAUUAUAUGCCA (((-(((.......)))))).((((..((((..(((((((((............)))))))))..)))).......(((((...(((....)))..)))))))))............... ( -24.90) >DroEre_CAF1 2053 104 + 1 -AGU------------UGGCUUGGCUCAAAGUUCUGACAGUGCAGUACAAAAUACACUGUGAGCCUCUUUAUUUCCGCUUAGAUAAGUUCACUUC---AGCGACACUAAAUUAUAUGCCA -...------------((((..(((((.........((((((............)))))))))))..........((((.....(((....))).---))))..............)))) ( -20.50) >DroYak_CAF1 11801 92 + 1 ----------------------------AAGUUUUGACAGUGCACUACAAAGUACACUGCUAGCCUUCUUAUUUCCGCUUAGAUAAGUUCACUUCAUAAGCGACACUAAAUUAUAUGCCA ----------------------------..(((..(.(((((.(((....))).)))))).)))...........((((((...(((....)))..)))))).................. ( -16.50) >consensus UGG_GU_UUUUU__UAUUCAUUGUCUUAAGAACUUCACAGUGCACUACAAAAUACACUGUGAGCCUCUUUAUUUCCGCUUAGAUAAGUUUACUUCAUAAGCGACACUAAAUUAUAUGCCA ...........................((((..(((((((((............)))))))))..))))......((((((...(((....)))..)))))).................. (-14.60 = -15.44 + 0.84)

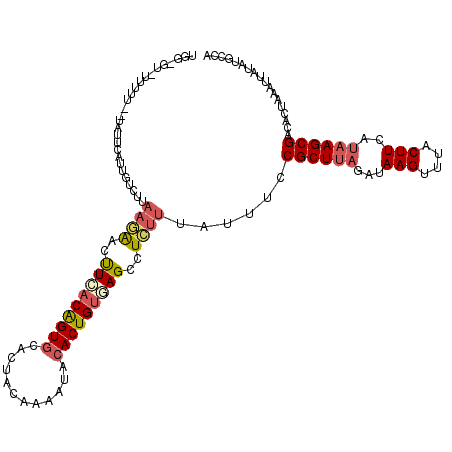
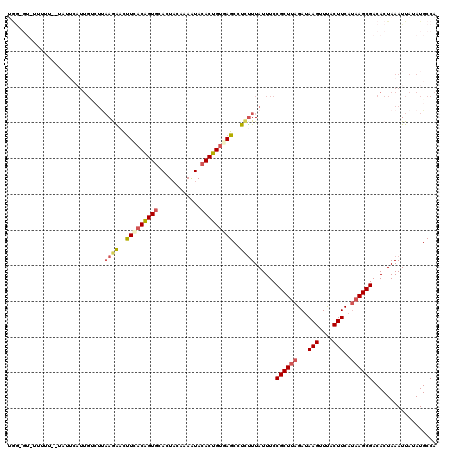
| Location | 3,162,457 – 3,162,577 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 77.83 |
| Mean single sequence MFE | -30.28 |
| Consensus MFE | -23.98 |
| Energy contribution | -24.50 |
| Covariance contribution | 0.52 |
| Combinations/Pair | 1.18 |
| Mean z-score | -3.16 |
| Structure conservation index | 0.79 |
| SVM decision value | 3.34 |
| SVM RNA-class probability | 0.999034 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
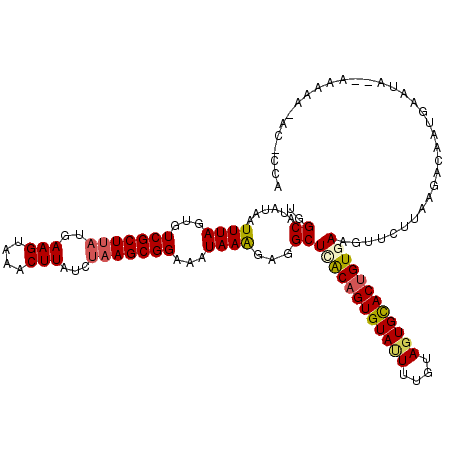
>3R_DroMel_CAF1 3162457 120 - 27905053 UGGCAUAUAAUUUAGUGUCGCUUAUGAAGUAAACUUAUCUAAGCGGAAAUAAAGAGGCUCACAGUGUAUUUUGUAGUGUACUGUGAAGUUCUUAAUGCAAUGAAUAAAAAAAAAACACCA ..((((......((...(((((((..(((....)))...)))))))...))(((((..((((((((((((....))))))))))))..))))).))))...................... ( -28.70) >DroSec_CAF1 10345 118 - 1 UGGCAUAUAAUUUAGUGUCGCUUAUGAAGUAAACUUAUCUAAGCGGAAAUAAAGAGGCUUACAGUGUAUUUUGAAGUGCACUGUGAAGUUCUUAAGACAAUGAAUAGGAAAAA-AC-CCA .........(((((.(((((((((..(((....)))...))))).......(((((..((((((((((((....))))))))))))..)))))..)))).))))).((.....-..-)). ( -32.10) >DroSim_CAF1 12513 119 - 1 UGGCAUAUAAUUUAGUGUCGCUUAUGAAGUAAACUUAUCUAAGCGGAAAUAAAGAGGCUCACAGUGUAUUUUGUAGUGCACCGUGAAGUUCUUAAGACAAUGAAUAGGAAAAAUAC-UCA .........(((((.(((((((((..(((....)))...))))).......(((((..((((.(((((((....))))))).))))..)))))..)))).)))))...........-... ( -29.40) >DroEre_CAF1 2053 104 - 1 UGGCAUAUAAUUUAGUGUCGCU---GAAGUGAACUUAUCUAAGCGGAAAUAAAGAGGCUCACAGUGUAUUUUGUACUGCACUGUCAGAACUUUGAGCCAAGCCA------------ACU- ((((......((((...(((((---.(((....))).....)))))...))))..(((((((((((((........)))))))).........)))))..))))------------...- ( -32.60) >DroYak_CAF1 11801 92 - 1 UGGCAUAUAAUUUAGUGUCGCUUAUGAAGUGAACUUAUCUAAGCGGAAAUAAGAAGGCUAGCAGUGUACUUUGUAGUGCACUGUCAAAACUU---------------------------- ((((......((((...(((((((..(((....)))...)))))))...))))...))))((((((((((....))))))))))........---------------------------- ( -28.60) >consensus UGGCAUAUAAUUUAGUGUCGCUUAUGAAGUAAACUUAUCUAAGCGGAAAUAAAGAGGCUCACAGUGUAUUUUGUAGUGCACUGUGAAGUUCUUAAGACAAUGAAUA__AAAAA_AC_CCA ..((......((((...(((((((..(((....)))...)))))))...))))...))((((((((((((....)))))))))))).................................. (-23.98 = -24.50 + 0.52)

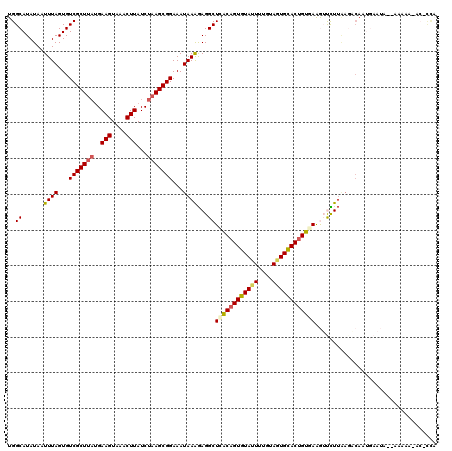
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:57:22 2006