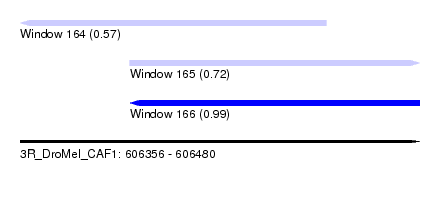
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 606,356 – 606,480 |
| Length | 124 |
| Max. P | 0.993041 |

| Location | 606,356 – 606,451 |
|---|---|
| Length | 95 |
| Sequences | 3 |
| Columns | 96 |
| Reading direction | reverse |
| Mean pairwise identity | 90.88 |
| Mean single sequence MFE | -19.43 |
| Consensus MFE | -18.23 |
| Energy contribution | -18.90 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.40 |
| Structure conservation index | 0.94 |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.565587 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 606356 95 - 27905053 CAAUAAGAGUACAUAUUAAUGUGUUUACUUUUUAUGGAGUGCUUAAA-AAAUGGAGCGACAAUUGCAGUAACCAACCAUAUUCCAAUUGGAUAUUG ((..(((((((.((((....)))).)))))))..))...(((((...-.....))))).((((((.((((........)))).))))))....... ( -15.70) >DroSec_CAF1 22434 93 - 1 CAAUAAGAGUAGAUUUUAGUGAGUUUACUCUUAAUGGAGUGCUUAAAAAAAUGGAGCUACAAUUGCAGUAACC---CAUAUUCCAAUUGGAUAUUG ((.(((((((((((((....))))))))))))).))....((((.........))))..((((((.((((...---..)))).))))))....... ( -22.40) >DroSim_CAF1 21591 93 - 1 CAAUAAGAGUAGAUUUUAGUGAGUUUACUCGUAAUGGAGUGCUUAAAAAAAUGGAGCUACAAUUGCAGUAACC---CAUUUUCCAAUUGGAUAUUG (((((.((((((((((....))))))))))(((((...((((((.........)))).)).))))).......---.....(((....)))))))) ( -20.20) >consensus CAAUAAGAGUAGAUUUUAGUGAGUUUACUCUUAAUGGAGUGCUUAAAAAAAUGGAGCUACAAUUGCAGUAACC___CAUAUUCCAAUUGGAUAUUG ((.(((((((((((((....))))))))))))).))....((((.........))))..((((((.((((........)))).))))))....... (-18.23 = -18.90 + 0.67)

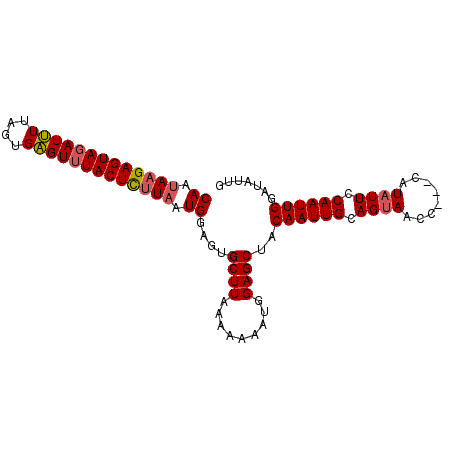
| Location | 606,390 – 606,480 |
|---|---|
| Length | 90 |
| Sequences | 3 |
| Columns | 91 |
| Reading direction | forward |
| Mean pairwise identity | 87.78 |
| Mean single sequence MFE | -14.53 |
| Consensus MFE | -13.11 |
| Energy contribution | -14.11 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.64 |
| Structure conservation index | 0.90 |
| SVM decision value | 0.41 |
| SVM RNA-class probability | 0.724423 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 606390 90 + 27905053 UUGUCGCUCCAUUU-UUUAAGCACUCCAUAAAAAGUAAACACAUUAAUAUGUACUCUUAUUGGAGAUUUCUUCUUUCGAUUUUAGUUUCGG .....(((......-....))).(((((.....(((....((((....))))))).....)))))...........(((........))). ( -11.50) >DroSec_CAF1 22465 87 + 1 UUGUAGCUCCAUUUUUUUAAGCACUCCAUUAAGAGUAAACUCACUAAAAUCUACUCUUAUUGGAGAUUUC---UUUUGAUUUUGGUUUCG- ........(((...((..(((..(((((.((((((((..............)))))))).)))))....)---))..))...))).....- ( -18.94) >DroSim_CAF1 21622 88 + 1 UUGUAGCUCCAUUUUUUUAAGCACUCCAUUACGAGUAAACUCACUAAAAUCUACUCUUAUUGGAGAUUUC---UUUCUAUUUUAGUUUCGU .....(((...........))).(((((.((.(((((..............))))).)).))))).....---.................. ( -13.14) >consensus UUGUAGCUCCAUUUUUUUAAGCACUCCAUUAAGAGUAAACUCACUAAAAUCUACUCUUAUUGGAGAUUUC___UUUCGAUUUUAGUUUCG_ .....(((...........))).(((((.((((((((..............)))))))).))))).......................... (-13.11 = -14.11 + 1.00)

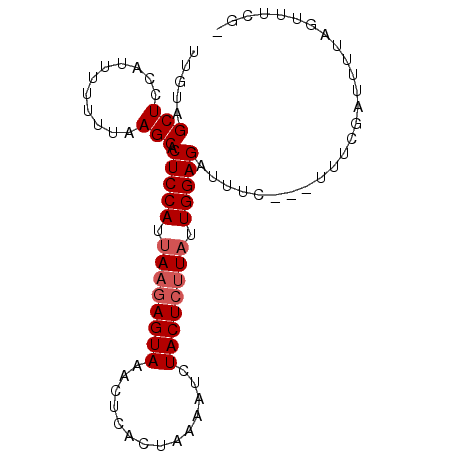
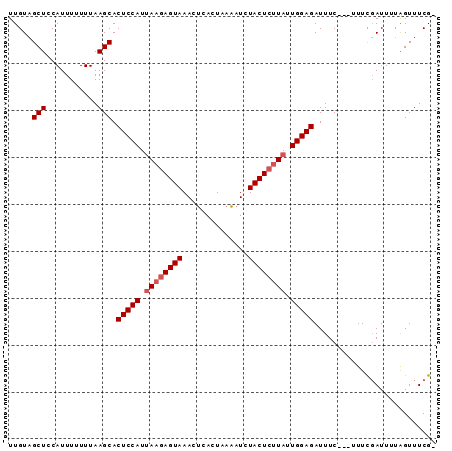
| Location | 606,390 – 606,480 |
|---|---|
| Length | 90 |
| Sequences | 3 |
| Columns | 91 |
| Reading direction | reverse |
| Mean pairwise identity | 87.78 |
| Mean single sequence MFE | -21.61 |
| Consensus MFE | -20.86 |
| Energy contribution | -21.20 |
| Covariance contribution | 0.34 |
| Combinations/Pair | 1.09 |
| Mean z-score | -4.06 |
| Structure conservation index | 0.97 |
| SVM decision value | 2.37 |
| SVM RNA-class probability | 0.993041 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
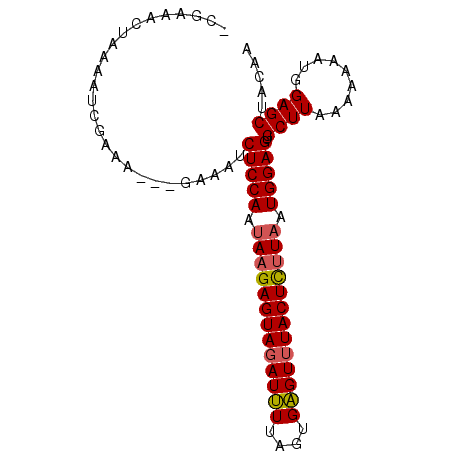
>3R_DroMel_CAF1 606390 90 - 27905053 CCGAAACUAAAAUCGAAAGAAGAAAUCUCCAAUAAGAGUACAUAUUAAUGUGUUUACUUUUUAUGGAGUGCUUAAA-AAAUGGAGCGACAA ............((....))......(((((..(((((((.((((....)))).)))))))..)))))(((((...-.....))))).... ( -19.70) >DroSec_CAF1 22465 87 - 1 -CGAAACCAAAAUCAAAA---GAAAUCUCCAAUAAGAGUAGAUUUUAGUGAGUUUACUCUUAAUGGAGUGCUUAAAAAAAUGGAGCUACAA -.....(((.......((---(....(((((.(((((((((((((....))))))))))))).)))))..))).......)))........ ( -25.44) >DroSim_CAF1 21622 88 - 1 ACGAAACUAAAAUAGAAA---GAAAUCUCCAAUAAGAGUAGAUUUUAGUGAGUUUACUCGUAAUGGAGUGCUUAAAAAAAUGGAGCUACAA ..................---.....(((((.((.((((((((((....)))))))))).)).))))).((((.........))))..... ( -19.70) >consensus _CGAAACUAAAAUCGAAA___GAAAUCUCCAAUAAGAGUAGAUUUUAGUGAGUUUACUCUUAAUGGAGUGCUUAAAAAAAUGGAGCUACAA ..........................(((((.(((((((((((((....))))))))))))).))))).((((.........))))..... (-20.86 = -21.20 + 0.34)

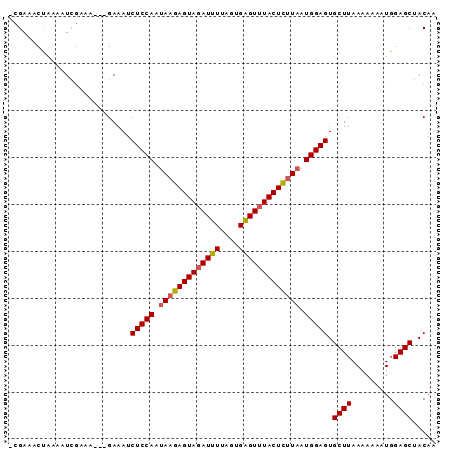
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:33:02 2006