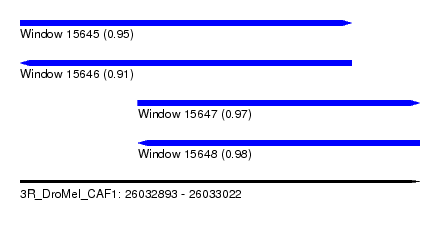
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 26,032,893 – 26,033,022 |
| Length | 129 |
| Max. P | 0.977044 |

| Location | 26,032,893 – 26,033,000 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 78.06 |
| Mean single sequence MFE | -34.62 |
| Consensus MFE | -23.44 |
| Energy contribution | -23.00 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.39 |
| Mean z-score | -2.06 |
| Structure conservation index | 0.68 |
| SVM decision value | 1.38 |
| SVM RNA-class probability | 0.947833 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 26032893 107 + 27905053 GUAAUGCAGUAGCCGUUGA--GAGUCGGCUCAACAUUGUGGAGUGAUUAGCGGC-----AGUUAAUCUACGUUCUGUUUGACAAUCGCUGAGCUUAGGU-AAUCGCUUAGCGAAU ((...((((.(((.(((((--(......))))))........(((((((((...-----.))))))).)))))))))...))..(((((((((......-....))))))))).. ( -36.20) >DroVir_CAF1 43654 112 + 1 GUUAUGCAGAAGCCUUGGAAUUGAUAGGCUCUGCAACGUGGAGUAAACUG--UUCUGGGUUUCAAUAUGCGUUCUGUUUGACAAUCGCUGAACAAGAAU-AAAAGUUCAGCGAAU ((((.((((((((.(((((((((((((((((..(...)..))))...)))--))...))))))))...)).)))))).))))..(((((((((......-....))))))))).. ( -38.50) >DroSec_CAF1 26015 107 + 1 GUAAUGCAGUAGCCGUGGA--GAGUCAGCUCAACAUUGUGGAGUGAUUAGCGGC-----AGUUAAUCUACGUUCUGUUUGACAAUCGCUGAGCUUAGGU-AAUCGCUUAGCGAAU ((...((((..(((((...--.(((((.(((.((...)).)))))))).)))))-----.((......))...))))...))..(((((((((......-....))))))))).. ( -34.00) >DroSim_CAF1 27647 107 + 1 GUAAUGCAGUAGCCGUGGA--GAGUCGGCUCAACAUUGUGGAGUGAUUAGCGGC-----AGUUAAUCUACGUUCUGUUUGACAAUCGCUGAGCUUAGGU-AAUCGCUUAGCGAAU ((...((((..(((((...--.(((((.(((.((...)).)))))))).)))))-----.((......))...))))...))..(((((((((......-....))))))))).. ( -33.60) >DroEre_CAF1 27084 107 + 1 GUAAUGCAGUAGCCGUGGC--AAGUCGGCUCAACAUUGUGGAGUGAUUAGCGGC-----CGUUAAUUUACGUUCUGUUUGACAAUCGCUGAGCUUGGGU-GAUCGCUUAGCGAAU ((...((((..(((((...--.(((((.(((.((...)).)))))))).)))))-----(((......)))..))))...))..(((((((((......-....))))))))).. ( -33.00) >DroAna_CAF1 28550 115 + 1 GUAAUGCUGUAGUCCUGGAUAAAGUCGACUAAGCAUUAUGGAGUGAUUAGCUGCCUGGUGAUUAUUCUGCGUUCUGUUUGACAAUCACUAGGGUAGGAUGAAACACCUAGCAAAU ((((((((.(((((((......))..)))))))))))))((.((..(((.(((((((((((((.((..((.....))..)).)))))))).)))))..))).)).))........ ( -32.40) >consensus GUAAUGCAGUAGCCGUGGA__GAGUCGGCUCAACAUUGUGGAGUGAUUAGCGGC_____AGUUAAUCUACGUUCUGUUUGACAAUCGCUGAGCUUAGGU_AAUCGCUUAGCGAAU ((...((((.(((((..........)))))......((((((.(((((...........))))).))))))..))))...))..(((((((((...........))))))))).. (-23.44 = -23.00 + -0.44)



| Location | 26,032,893 – 26,033,000 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 78.06 |
| Mean single sequence MFE | -25.02 |
| Consensus MFE | -18.14 |
| Energy contribution | -17.12 |
| Covariance contribution | -1.02 |
| Combinations/Pair | 1.44 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.72 |
| SVM decision value | 1.04 |
| SVM RNA-class probability | 0.905913 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 26032893 107 - 27905053 AUUCGCUAAGCGAUU-ACCUAAGCUCAGCGAUUGUCAAACAGAACGUAGAUUAACU-----GCCGCUAAUCACUCCACAAUGUUGAGCCGACUC--UCAACGGCUACUGCAUUAC ..(((((.(((....-......))).)))))..............((.(((((...-----.....))))))).....(((((..(((((....--....)))))...))))).. ( -24.60) >DroVir_CAF1 43654 112 - 1 AUUCGCUGAACUUUU-AUUCUUGUUCAGCGAUUGUCAAACAGAACGCAUAUUGAAACCCAGAA--CAGUUUACUCCACGUUGCAGAGCCUAUCAAUUCCAAGGCUUCUGCAUAAC ..(((((((((....-......)))))))))...........((((.......((((......--..))))......))))(((((((((..........)))).)))))..... ( -29.12) >DroSec_CAF1 26015 107 - 1 AUUCGCUAAGCGAUU-ACCUAAGCUCAGCGAUUGUCAAACAGAACGUAGAUUAACU-----GCCGCUAAUCACUCCACAAUGUUGAGCUGACUC--UCCACGGCUACUGCAUUAC ..(((((.(((....-......))).)))))..............((.(((((...-----.....))))))).....(((((..(((((....--....)))))...))))).. ( -22.40) >DroSim_CAF1 27647 107 - 1 AUUCGCUAAGCGAUU-ACCUAAGCUCAGCGAUUGUCAAACAGAACGUAGAUUAACU-----GCCGCUAAUCACUCCACAAUGUUGAGCCGACUC--UCCACGGCUACUGCAUUAC ..(((((.(((....-......))).)))))..............((.(((((...-----.....))))))).....(((((..(((((....--....)))))...))))).. ( -24.60) >DroEre_CAF1 27084 107 - 1 AUUCGCUAAGCGAUC-ACCCAAGCUCAGCGAUUGUCAAACAGAACGUAAAUUAACG-----GCCGCUAAUCACUCCACAAUGUUGAGCCGACUU--GCCACGGCUACUGCAUUAC ..(((((.(((....-......))).)))))..........(..(((......)))-----..)..............(((((..(((((....--....)))))...))))).. ( -24.40) >DroAna_CAF1 28550 115 - 1 AUUUGCUAGGUGUUUCAUCCUACCCUAGUGAUUGUCAAACAGAACGCAGAAUAAUCACCAGGCAGCUAAUCACUCCAUAAUGCUUAGUCGACUUUAUCCAGGACUACAGCAUUAC ....((((((........)))..(((.(((((((((............).)))))))).))).)))...........((((((((((((............))))).))))))). ( -25.00) >consensus AUUCGCUAAGCGAUU_ACCCAAGCUCAGCGAUUGUCAAACAGAACGUAGAUUAACU_____GCCGCUAAUCACUCCACAAUGUUGAGCCGACUC__UCCACGGCUACUGCAUUAC ..(((((.(((...........))).)))))...............................................(((((..(((((..........)))))...))))).. (-18.14 = -17.12 + -1.02)


| Location | 26,032,931 – 26,033,022 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 75.80 |
| Mean single sequence MFE | -25.48 |
| Consensus MFE | -19.26 |
| Energy contribution | -18.54 |
| Covariance contribution | -0.72 |
| Combinations/Pair | 1.30 |
| Mean z-score | -1.78 |
| Structure conservation index | 0.76 |
| SVM decision value | 1.63 |
| SVM RNA-class probability | 0.969008 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 26032931 91 + 27905053 GAGUGAUUAGCGGC-----AGUUAAUCUACGUUCUGUUUGACAAUCGCUGAGCU--UAGGU-AAUCGCUUAGCGAAUAACACAAGAACGGAGUUCACAA (((.(((((((...-----.)))))))..((((((...((....(((((((((.--.....-....)))))))))....))..))))))...))).... ( -28.30) >DroPse_CAF1 29834 96 + 1 GAGUGAGUAGUUGCUUUGAGUUUACUGUACGUUCUGUUUGACAAUCACAGGACUAUAAGGU-GA--GUCCAGUGAAUAACACAAGAACGGCGGAUAUAA (((..(....)..)))........((((.((((((...((....((((.(((((.......-.)--)))).))))....))..))))))))))...... ( -23.90) >DroSim_CAF1 27685 91 + 1 GAGUGAUUAGCGGC-----AGUUAAUCUACGUUCUGUUUGACAAUCGCUGAGCU--UAGGU-AAUCGCUUAGCGAAUAACACAAGAACGGAGUUCACAA (((.(((((((...-----.)))))))..((((((...((....(((((((((.--.....-....)))))))))....))..))))))...))).... ( -28.30) >DroEre_CAF1 27122 91 + 1 GAGUGAUUAGCGGC-----CGUUAAUUUACGUUCUGUUUGACAAUCGCUGAGCU--UGGGU-GAUCGCUUAGCGAAUAACACAAGAACGGUAUUUACAA ..(((((((((...-----.)))))....((((((...((....(((((((((.--.....-....)))))))))....))..))))))....)))).. ( -25.50) >DroAna_CAF1 28590 93 + 1 GAGUGAUUAGCUGCCUGGUGAUUAUUCUGCGUUCUGUUUGACAAUCACUAGGGU--AGGAUGAAACACCUAGCAAAUAACACAAGAACGGU----ACAG ..(((.(((..(((..((((.(((((((((..((((..((.....)).))))))--)))))))..))))..)))..)))))).........----.... ( -23.00) >DroPer_CAF1 30482 96 + 1 GAGUGAGUAGUUGCUUUGAGUUUACUGUACGUUCUGUUUGACAAUCACAGGACUAUAAGGU-GA--GUCCAGUGAAUAACACAAGAACGGCGGAUAUAA (((..(....)..)))........((((.((((((...((....((((.(((((.......-.)--)))).))))....))..))))))))))...... ( -23.90) >consensus GAGUGAUUAGCGGC_____GGUUAAUCUACGUUCUGUUUGACAAUCACUGAGCU__AAGGU_AAUCGCCUAGCGAAUAACACAAGAACGGAGUUUACAA ....((((((...........))))))..((((((...((....(((((((((.............)))))))))....))..)))))).......... (-19.26 = -18.54 + -0.72)



| Location | 26,032,931 – 26,033,022 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 75.80 |
| Mean single sequence MFE | -21.20 |
| Consensus MFE | -16.19 |
| Energy contribution | -15.14 |
| Covariance contribution | -1.05 |
| Combinations/Pair | 1.28 |
| Mean z-score | -1.92 |
| Structure conservation index | 0.76 |
| SVM decision value | 1.78 |
| SVM RNA-class probability | 0.977044 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 26032931 91 - 27905053 UUGUGAACUCCGUUCUUGUGUUAUUCGCUAAGCGAUU-ACCUA--AGCUCAGCGAUUGUCAAACAGAACGUAGAUUAACU-----GCCGCUAAUCACUC ..((((.....(((((..((.((.(((((.(((....-.....--.))).))))).)).))...)))))((((.....))-----))......)))).. ( -23.70) >DroPse_CAF1 29834 96 - 1 UUAUAUCCGCCGUUCUUGUGUUAUUCACUGGAC--UC-ACCUUAUAGUCCUGUGAUUGUCAAACAGAACGUACAGUAAACUCAAAGCAACUACUCACUC ........((((((((..((.((.((((.((((--(.-.......))))).)))).)).))...))))))...((....))....))............ ( -19.90) >DroSim_CAF1 27685 91 - 1 UUGUGAACUCCGUUCUUGUGUUAUUCGCUAAGCGAUU-ACCUA--AGCUCAGCGAUUGUCAAACAGAACGUAGAUUAACU-----GCCGCUAAUCACUC ..((((.....(((((..((.((.(((((.(((....-.....--.))).))))).)).))...)))))((((.....))-----))......)))).. ( -23.70) >DroEre_CAF1 27122 91 - 1 UUGUAAAUACCGUUCUUGUGUUAUUCGCUAAGCGAUC-ACCCA--AGCUCAGCGAUUGUCAAACAGAACGUAAAUUAACG-----GCCGCUAAUCACUC .(((.(((..((((((..((.((.(((((.(((....-.....--.))).))))).)).))...))))))...))).)))-----.............. ( -19.40) >DroAna_CAF1 28590 93 - 1 CUGU----ACCGUUCUUGUGUUAUUUGCUAGGUGUUUCAUCCU--ACCCUAGUGAUUGUCAAACAGAACGCAGAAUAAUCACCAGGCAGCUAAUCACUC ((((----..((((((..((.((.(..(((((.((........--)))))))..).)).))...))))))..((....)).....)))).......... ( -20.60) >DroPer_CAF1 30482 96 - 1 UUAUAUCCGCCGUUCUUGUGUUAUUCACUGGAC--UC-ACCUUAUAGUCCUGUGAUUGUCAAACAGAACGUACAGUAAACUCAAAGCAACUACUCACUC ........((((((((..((.((.((((.((((--(.-.......))))).)))).)).))...))))))...((....))....))............ ( -19.90) >consensus UUGUAAACACCGUUCUUGUGUUAUUCGCUAAGCGAUC_ACCUA__AGCCCAGCGAUUGUCAAACAGAACGUAGAUUAACC_____GCAGCUAAUCACUC ..........((((((..((.((.(((((.(((.............))).))))).)).))...))))))............................. (-16.19 = -15.14 + -1.05)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:42:25 2006