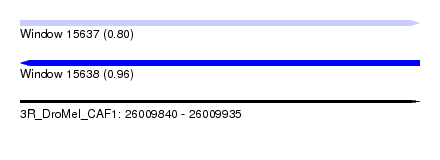
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 26,009,840 – 26,009,935 |
| Length | 95 |
| Max. P | 0.956053 |

| Location | 26,009,840 – 26,009,935 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 101 |
| Reading direction | forward |
| Mean pairwise identity | 80.48 |
| Mean single sequence MFE | -30.03 |
| Consensus MFE | -18.70 |
| Energy contribution | -18.90 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.61 |
| SVM RNA-class probability | 0.800304 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 26009840 95 + 27905053 AAGAUCGGCGUACGUGCCGAGUGCACGUAGGCCAUGCCAAGUGGAGGGAGUCAAGGG------GCAGGUGGGGAAGUUAGUUAAUUGAUUAGGGAACUAUG ..(((((((.(((((((.....))))))).))).((((...(((......)))...)------)))....................))))((....))... ( -29.10) >DroSec_CAF1 3745 95 + 1 AAGAUCGGCGUACGUGCCGAGUGCACGUAGGCCAUGCCAAGUGGAGGGAGUCGAGGG------GCAGGUGGGGAAGUUAGUUAAUUGAUUAGGGAACUAUG ..(((((((.(((((((.....))))))).))).((((...(((......)))...)------)))....................))))((....))... ( -28.40) >DroSim_CAF1 3852 95 + 1 AAGAUCGGCGUACGUGCCGAGUGCACGUAGGCCAUGCCAAGUGGAGGGUGUCAAGGG------GCAGGUGGGGAAGUUAGUUAAUUGAUUAGGGAACUAUG ..(((((((.(((((((.....))))))).))).((((...(((......)))...)------)))....................))))((....))... ( -29.10) >DroEre_CAF1 3955 95 + 1 GAGAUCGGUGUACGUGCCGCGUGCACGUAGGCCAGGCCAAGUGGAGGAUCUUGGGGG------CUAGGCGGGGAAGUUAGUUAAUUGAUUAGGGAACUAUG (((((((((.(((((((.....))))))).)))...((....))..))))))...((------((((.(......))))))).......(((....))).. ( -29.50) >DroYak_CAF1 3819 101 + 1 AAGAUCGGCGUACGUGCCGAGUGCACGUAGGCCAGACCAAGUGGAGGAUCCCAGGGGGGGAUCUCAGGUGGGGAAGUUAGUUAAUUGAUUAGGGAACUAUG ..(((((((.(((((((.....))))))).)))...(((..((..(((((((.....)))))))))..)))...............))))((....))... ( -37.30) >DroAna_CAF1 4310 84 + 1 ------GGCGUACGUGCCAAGUGCACGUAGGCCUCGUCAGAUUCUUGGAGCUCAGGG------GCAAGUGGUGGAG-----CAAUCAAUUAAGGAACAAUU ------(((.(((((((.....))))))).)))......(.(((((((.(((....)------))...((((....-----..)))).))))))).).... ( -26.80) >consensus AAGAUCGGCGUACGUGCCGAGUGCACGUAGGCCAUGCCAAGUGGAGGGAGUCAAGGG______GCAGGUGGGGAAGUUAGUUAAUUGAUUAGGGAACUAUG ......(((.(((((((.....))))))).)))........................................................(((....))).. (-18.70 = -18.90 + 0.20)


| Location | 26,009,840 – 26,009,935 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 101 |
| Reading direction | reverse |
| Mean pairwise identity | 80.48 |
| Mean single sequence MFE | -22.64 |
| Consensus MFE | -15.62 |
| Energy contribution | -15.78 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.98 |
| Structure conservation index | 0.69 |
| SVM decision value | 1.46 |
| SVM RNA-class probability | 0.956053 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 26009840 95 - 27905053 CAUAGUUCCCUAAUCAAUUAACUAACUUCCCCACCUGC------CCCUUGACUCCCUCCACUUGGCAUGGCCUACGUGCACUCGGCACGUACGCCGAUCUU ............(((....................(((------(...((........))...)))).(((.(((((((.....))))))).))))))... ( -21.80) >DroSec_CAF1 3745 95 - 1 CAUAGUUCCCUAAUCAAUUAACUAACUUCCCCACCUGC------CCCUCGACUCCCUCCACUUGGCAUGGCCUACGUGCACUCGGCACGUACGCCGAUCUU ............(((....................(((------(....((.....)).....)))).(((.(((((((.....))))))).))))))... ( -20.60) >DroSim_CAF1 3852 95 - 1 CAUAGUUCCCUAAUCAAUUAACUAACUUCCCCACCUGC------CCCUUGACACCCUCCACUUGGCAUGGCCUACGUGCACUCGGCACGUACGCCGAUCUU ............(((....................(((------(...((........))...)))).(((.(((((((.....))))))).))))))... ( -21.80) >DroEre_CAF1 3955 95 - 1 CAUAGUUCCCUAAUCAAUUAACUAACUUCCCCGCCUAG------CCCCCAAGAUCCUCCACUUGGCCUGGCCUACGUGCACGCGGCACGUACACCGAUCUC ............(((.................(((.((------...(((((........))))).))))).(((((((.....)))))))....)))... ( -19.70) >DroYak_CAF1 3819 101 - 1 CAUAGUUCCCUAAUCAAUUAACUAACUUCCCCACCUGAGAUCCCCCCCUGGGAUCCUCCACUUGGUCUGGCCUACGUGCACUCGGCACGUACGCCGAUCUU ..(((((............)))))............((((((((.....))))).))).....((((.(((.(((((((.....))))))).))))))).. ( -29.70) >DroAna_CAF1 4310 84 - 1 AAUUGUUCCUUAAUUGAUUG-----CUCCACCACUUGC------CCCUGAGCUCCAAGAAUCUGACGAGGCCUACGUGCACUUGGCACGUACGCC------ ..(((((......(((...(-----(((..........------....))))..)))......)))))(((.(((((((.....))))))).)))------ ( -22.24) >consensus CAUAGUUCCCUAAUCAAUUAACUAACUUCCCCACCUGC______CCCUUGACACCCUCCACUUGGCAUGGCCUACGUGCACUCGGCACGUACGCCGAUCUU ....................................................................(((.(((((((.....))))))).)))...... (-15.62 = -15.78 + 0.17)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:42:16 2006