| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 25,957,485 – 25,957,595 |
| Length | 110 |
| Max. P | 0.880408 |

| Location | 25,957,485 – 25,957,595 |
|---|---|
| Length | 110 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 76.67 |
| Mean single sequence MFE | -29.62 |
| Consensus MFE | -18.72 |
| Energy contribution | -19.25 |
| Covariance contribution | 0.53 |
| Combinations/Pair | 1.32 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.67 |
| SVM RNA-class probability | 0.818465 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
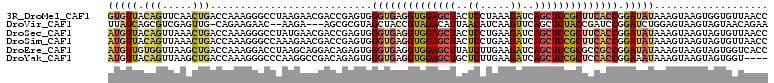
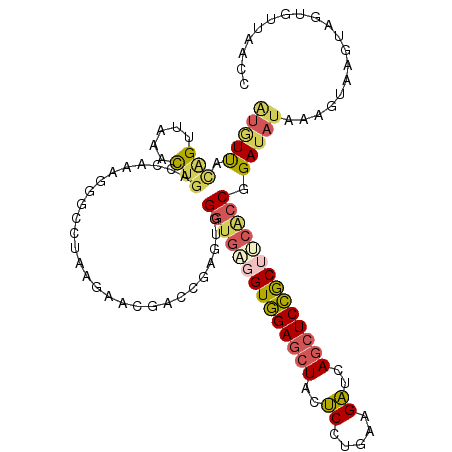
>3R_DroMel_CAF1 25957485 110 + 27905053 GGUUAACACCACUUACUUUAUAUCCGGUGAAGCGGAGCUGAUCUUUAGGAGUAGCUCCACCUCACCCACUCGGUCGUUCUUAGGCCCUUUGGUCAGUUGAACUGUAACAC .((((..((((..............(((((.(.(((((((.((.....)).))))))).).))))).....((((.......))))...))))(((.....))))))).. ( -32.60) >DroVir_CAF1 14873 104 + 1 UUCUGUUACUACUUACUCCAGAUCCGGAUCGGUAGAGCUGAACUUGAUGUUAAUGUCUACGGUAGCUACGCGCU---UCUU--GUUCUUCUG-CAACUCGACGCUGAUAA (((.(((.(((((...(((......)))..)))))))).))).....(((((.((((...((.(((.....)))---))((--((......)-)))...)))).))))). ( -19.70) >DroSec_CAF1 9686 110 + 1 GGUUAACACUACUUACUUUAUAUCCGGUGAAGCGGAGCUGAUCUUCAGGAGUAGCUCCACCUCACCCACUCGGUCGUUCAUAGGCCCUUUGGUCAGUUUAACUGUAACAU ((((((.((((((............(((((.(.(((((((.((.....)).))))))).).))))).....((((.......))))....))).)))))))))....... ( -32.30) >DroSim_CAF1 9674 110 + 1 GGUUAACACUACUUACUUUAUAUCCGGUGAAGCGGAGCUGAUCUUCAGGAGUAGCUCCACCUCACCCACUCGGUCGUUCUUUGGCCCUUUGGUCAGUUUAACUGUAACAU ((((((.((((((............(((((.(.(((((((.((.....)).))))))).).))))).....(((((.....)))))....))).)))))))))....... ( -32.90) >DroEre_CAF1 7692 110 + 1 GGUGACCACUACUUACUUUAUAUCCGGCGGCGCGGAGCUGAUCUUCAAGAUAAGCUCCACCUCACCCACUCUGUCCUGCUUAGGUCCUUUGGUCAGCUUAACCACAACAU (.((((((.................((.((.(.((((((.(((.....))).)))))).).)).))......(.(((....))).)...)))))).)............. ( -28.20) >DroYak_CAF1 7773 106 + 1 ----ACCACUACUUACUUUAUUUCCGGUGGAGCGGAGCUGAUCUUCAAGAGCAGCUCCACCUCACCCACUCUGUCGGCCUUGGGCCCUUUGGUCAGCUUAACUGUAACAU ----((((.................((((..(.(((((((.((.....)).))))))).)..)))).........((((...))))...))))(((.....)))...... ( -32.00) >consensus GGUUAACACUACUUACUUUAUAUCCGGUGAAGCGGAGCUGAUCUUCAGGAGUAGCUCCACCUCACCCACUCGGUCGUUCUUAGGCCCUUUGGUCAGCUUAACUGUAACAU .........................(((((.(.(((((((.((.....)).))))))).).)))))................((((....))))................ (-18.72 = -19.25 + 0.53)

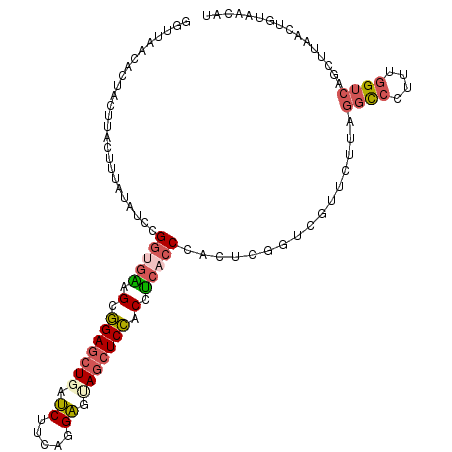
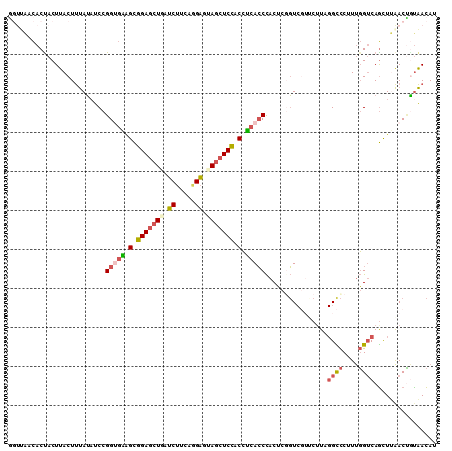
| Location | 25,957,485 – 25,957,595 |
|---|---|
| Length | 110 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 76.67 |
| Mean single sequence MFE | -32.66 |
| Consensus MFE | -22.57 |
| Energy contribution | -23.22 |
| Covariance contribution | 0.65 |
| Combinations/Pair | 1.46 |
| Mean z-score | -1.50 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.91 |
| SVM RNA-class probability | 0.880408 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 25957485 110 - 27905053 GUGUUACAGUUCAACUGACCAAAGGGCCUAAGAACGACCGAGUGGGUGAGGUGGAGCUACUCCUAAAGAUCAGCUCCGCUUCACCGGAUAUAAAGUAAGUGGUGUUAACC ..(((((((.....)))((((..((............)).....((((((((((((((..((.....))..))))))))))))))..............))))..)))). ( -34.80) >DroVir_CAF1 14873 104 - 1 UUAUCAGCGUCGAGUUG-CAGAAGAAC--AAGA---AGCGCGUAGCUACCGUAGACAUUAACAUCAAGUUCAGCUCUACCGAUCCGGAUCUGGAGUAAGUAGUAACAGAA ...((.((((....(((-........)--))..---.))))((.(((((.(((((....(((.....)))....)))))...((((....))))....))))).)).)). ( -19.50) >DroSec_CAF1 9686 110 - 1 AUGUUACAGUUAAACUGACCAAAGGGCCUAUGAACGACCGAGUGGGUGAGGUGGAGCUACUCCUGAAGAUCAGCUCCGCUUCACCGGAUAUAAAGUAAGUAGUGUUAACC ......(((.....))).......(((((((..((.......(.((((((((((((((..((.....))..)))))))))))))).).......))..)))).))).... ( -35.34) >DroSim_CAF1 9674 110 - 1 AUGUUACAGUUAAACUGACCAAAGGGCCAAAGAACGACCGAGUGGGUGAGGUGGAGCUACUCCUGAAGAUCAGCUCCGCUUCACCGGAUAUAAAGUAAGUAGUGUUAACC ........(((((((((..((..((............))...))((((((((((((((..((.....))..))))))))))))))..............)))).))))). ( -33.40) >DroEre_CAF1 7692 110 - 1 AUGUUGUGGUUAAGCUGACCAAAGGACCUAAGCAGGACAGAGUGGGUGAGGUGGAGCUUAUCUUGAAGAUCAGCUCCGCGCCGCCGGAUAUAAAGUAAGUAGUGGUCACC ......(((((.....)))))...((((...((...((....(.((((..((((((((.(((.....))).))))))))..)))).).......))..))...))))... ( -33.50) >DroYak_CAF1 7773 106 - 1 AUGUUACAGUUAAGCUGACCAAAGGGCCCAAGGCCGACAGAGUGGGUGAGGUGGAGCUGCUCUUGAAGAUCAGCUCCGCUCCACCGGAAAUAAAGUAAGUAGUGGU---- ......(((.....)))((((...((((...)))).........((((..(((((((((.((.....)).)))))))))..)))).................))))---- ( -39.40) >consensus AUGUUACAGUUAAACUGACCAAAGGGCCUAAGAACGACCGAGUGGGUGAGGUGGAGCUACUCCUGAAGAUCAGCUCCGCUUCACCGGAUAUAAAGUAAGUAGUGUUAACC (((((.(((.....)))...........................((((((((((((((..((.....))..)))))))))))))).)))))................... (-22.57 = -23.22 + 0.65)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:42:04 2006