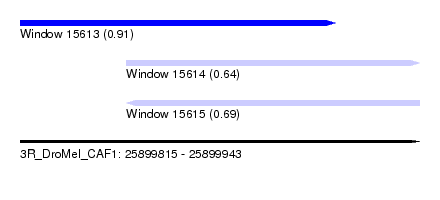
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 25,899,815 – 25,899,943 |
| Length | 128 |
| Max. P | 0.912984 |

| Location | 25,899,815 – 25,899,916 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 75.68 |
| Mean single sequence MFE | -32.02 |
| Consensus MFE | -13.70 |
| Energy contribution | -14.10 |
| Covariance contribution | 0.39 |
| Combinations/Pair | 1.20 |
| Mean z-score | -2.37 |
| Structure conservation index | 0.43 |
| SVM decision value | 1.09 |
| SVM RNA-class probability | 0.912984 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 25899815 101 + 27905053 UGUCCUCUUUAUCGGCGACUUUGGGUUGCAUACUUUUGUGCUCUUGGGUGGGU---AGC-AAAGUUUUGCCGCCUUUGCAAGAUUUU-----CGCAAGUCUUGCAGAAAA ............((((((..((..(((((.(((((..(....)..))))).))---)))-..))..))))))..((((((((((((.-----...))))))))))))... ( -31.10) >DroVir_CAF1 42920 90 + 1 CU--UUUGCUAUCAGCGUCUU-----UGCAUACUUUUCUGCUCUCAGUCA-G----UUG--AAACUUUGCCGU-UUUGCAAGUUUUU-----CGCAAGUCUUGCAGAAAA ..--.......(((((..((.-----.(((........)))....))...-)----)))--)..........(-((((((((.....-----(....).))))))))).. ( -17.90) >DroPse_CAF1 28180 99 + 1 UUUGGCCUUUAUCGGGGGCCU-----UGCAUACUUUUGUGCGCUGCCGCUGGUGGAAUG-AAAGUUUUGCCGCCUCUGCAAGUUUUU-----CGCAAGUCUUGCAGAAAA ...(((.......((.(((..-----((((((....))))))..))).))((..((((.-...))))..)))))((((((((.....-----(....).))))))))... ( -39.70) >DroYak_CAF1 34403 102 + 1 UGUCCUCUUUAUCGGUGGCUUUGGCUUGCAUACUUUUGUGCGCUGGGGUGGGU---GGGCAAAGUUUCGCCGCCUUUGCAAGUUUUU-----CGCAAGUCUUGCAGAAAA .............((((((....(((..(.(((((..(....)..))))).).---.)))........))))))((((((((.....-----(....).))))))))... ( -32.20) >DroAna_CAF1 26988 100 + 1 UGUUCGCUUUAUCGGGGGCCU-----UGCAUACUUUUGUGCUUCGGGAU-GGC---AGA-AAAGUUUUGCCGCCUUGGCAAGUUUUUUUUUCUGCAAGUCUUGCAGAAAA .....((((...(.(((((..-----.(((.((((((.((((.......-)))---).)-)))))..))).))))).).))))....(((((((((.....))))))))) ( -31.50) >DroPer_CAF1 27984 99 + 1 UUUGGCCUUUAUCGGGGGCCU-----UGCAUACUUUUGUGCGCUGCCGCUGGUGGAAUG-AAAGUUUUGCCGCCUCUGCAAGUUUUU-----CGCAAGUCUUGCAGAAAA ...(((.......((.(((..-----((((((....))))))..))).))((..((((.-...))))..)))))((((((((.....-----(....).))))))))... ( -39.70) >consensus UGUCCUCUUUAUCGGGGGCCU_____UGCAUACUUUUGUGCGCUGGGGCGGGU___AGG_AAAGUUUUGCCGCCUUUGCAAGUUUUU_____CGCAAGUCUUGCAGAAAA .............((.(((........(((((....))))).((......))................))).))((((((((.................))))))))... (-13.70 = -14.10 + 0.39)

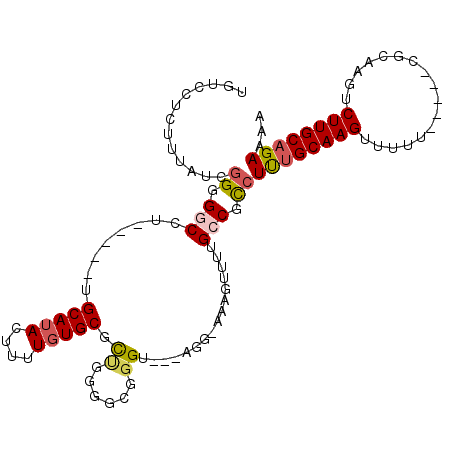
| Location | 25,899,849 – 25,899,943 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 82.57 |
| Mean single sequence MFE | -29.79 |
| Consensus MFE | -16.28 |
| Energy contribution | -17.26 |
| Covariance contribution | 0.98 |
| Combinations/Pair | 1.29 |
| Mean z-score | -2.69 |
| Structure conservation index | 0.55 |
| SVM decision value | 0.21 |
| SVM RNA-class probability | 0.635274 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 25899849 94 + 27905053 UUUGUGCUCUUGGGUGGGUAGC-AAAGUUUUGCCGCCUUUGCAAGAUUUU-----CGCAAGUCUUGCAGAAAAUUAUACAAAAGGCGAAGGCAAAGCUUA ..(((((((......))))).)-)(((((((((((((((((((((((((.-----...)))))))))).............)))))...)))))))))). ( -32.61) >DroVir_CAF1 42947 90 + 1 UUUCUGCUCUCAGUCA-G-UUG--AAACUUUGCCGU-UUUGCAAGUUUUU-----CGCAAGUCUUGCAGAAAAUUAUACAAAAGGCAAACGUGUUGUGUG .(((.(((.......)-)-).)--))((((((((.(-((((((((.....-----(....).)))))))))............)))))).))........ ( -19.22) >DroSim_CAF1 27142 94 + 1 UUUGUGCUCUUGGGUGGGUAGC-AAAGUUUUGCCGCCUUUGCAAGAUUUU-----CGCAAGUCUUGCAGAAAAUUAUACAAAAGGCGAAGGCAAAGCUUA ..(((((((......))))).)-)(((((((((((((((((((((((((.-----...)))))))))).............)))))...)))))))))). ( -32.61) >DroEre_CAF1 35156 94 + 1 UUUGUGCGCUUGGGUGGGAGGG-AAAGUUUCGCCGCCUUUGCAAGUUUUU-----CGCAAGUCUUGCAGAAAAUUAUACAAAAGGCGAAGGCAAAGCUUA .......((((.((((..(...-....)..))))((((((((.((((((.-----.(((.....)))..))))))...(....))))))))).))))... ( -28.90) >DroYak_CAF1 34437 95 + 1 UUUGUGCGCUGGGGUGGGUGGGCAAAGUUUCGCCGCCUUUGCAAGUUUUU-----CGCAAGUCUUGCAGAAAAUUAUACAAAAGGCGAAGGCGAAGCUUA ..(((.((((......)))).)))((((((((((((((((...((((((.-----.(((.....)))..)))))).....))))))...)))))))))). ( -35.10) >DroAna_CAF1 27017 94 + 1 UUUGUGCUUCGGGAU-GGCAGA-AAAGUUUUGCCGCCUUGGCAAGUUUUUUUUUCUGCAAGUCUUGCAGAAAAUUAUACAAAAGGCGAA----AAGCUUA ....((((..(((.(-((((((-.....)))))))))).)))).((.(..(((((((((.....)))))))))..).))...((((...----..)))). ( -30.30) >consensus UUUGUGCUCUUGGGUGGGUAGG_AAAGUUUUGCCGCCUUUGCAAGUUUUU_____CGCAAGUCUUGCAGAAAAUUAUACAAAAGGCGAAGGCAAAGCUUA ........................(((((((((((((((((((((.................)))))).............)))))...)))))))))). (-16.28 = -17.26 + 0.98)

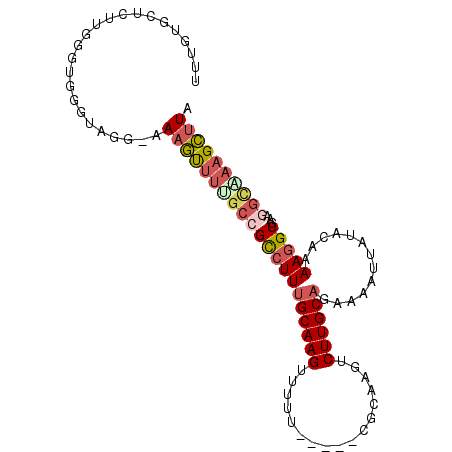

| Location | 25,899,849 – 25,899,943 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 82.57 |
| Mean single sequence MFE | -24.90 |
| Consensus MFE | -12.06 |
| Energy contribution | -13.15 |
| Covariance contribution | 1.09 |
| Combinations/Pair | 1.19 |
| Mean z-score | -3.18 |
| Structure conservation index | 0.48 |
| SVM decision value | 0.34 |
| SVM RNA-class probability | 0.694563 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 25899849 94 - 27905053 UAAGCUUUGCCUUCGCCUUUUGUAUAAUUUUCUGCAAGACUUGCG-----AAAAUCUUGCAAAGGCGGCAAAACUUU-GCUACCCACCCAAGAGCACAAA ...(((((.....(((((((.(((..((((((.(((.....))))-----)))))..))))))))))((((....))-))..........)))))..... ( -27.50) >DroVir_CAF1 42947 90 - 1 CACACAACACGUUUGCCUUUUGUAUAAUUUUCUGCAAGACUUGCG-----AAAAACUUGCAAA-ACGGCAAAGUUU--CAA-C-UGACUGAGAGCAGAAA ...........(((((((((((((...(((((.(((.....))))-----))))...))))))-).)))))).(((--((.-.-....)))))....... ( -20.90) >DroSim_CAF1 27142 94 - 1 UAAGCUUUGCCUUCGCCUUUUGUAUAAUUUUCUGCAAGACUUGCG-----AAAAUCUUGCAAAGGCGGCAAAACUUU-GCUACCCACCCAAGAGCACAAA ...(((((.....(((((((.(((..((((((.(((.....))))-----)))))..))))))))))((((....))-))..........)))))..... ( -27.50) >DroEre_CAF1 35156 94 - 1 UAAGCUUUGCCUUCGCCUUUUGUAUAAUUUUCUGCAAGACUUGCG-----AAAAACUUGCAAAGGCGGCGAAACUUU-CCCUCCCACCCAAGCGCACAAA .(((.((((((...((((((.(((...(((((.(((.....))))-----))))...))))))))))))))).))).-...................... ( -22.40) >DroYak_CAF1 34437 95 - 1 UAAGCUUCGCCUUCGCCUUUUGUAUAAUUUUCUGCAAGACUUGCG-----AAAAACUUGCAAAGGCGGCGAAACUUUGCCCACCCACCCCAGCGCACAAA ...((((((((...((((((.(((...(((((.(((.....))))-----))))...))))))))))))))).....((............))))..... ( -24.80) >DroAna_CAF1 27017 94 - 1 UAAGCUU----UUCGCCUUUUGUAUAAUUUUCUGCAAGACUUGCAGAAAAAAAAACUUGCCAAGGCGGCAAAACUUU-UCUGCC-AUCCCGAAGCACAAA ...((((----(..(((((..(((...(((((((((.....))))))))).......))).)))))((((.......-..))))-.....)))))..... ( -26.30) >consensus UAAGCUUUGCCUUCGCCUUUUGUAUAAUUUUCUGCAAGACUUGCG_____AAAAACUUGCAAAGGCGGCAAAACUUU_CCUACCCACCCAAGAGCACAAA .(((.(((((...(((((((.(((...((((.((((.....)))).....))))...))))))))))))))).)))........................ (-12.06 = -13.15 + 1.09)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:41:55 2006