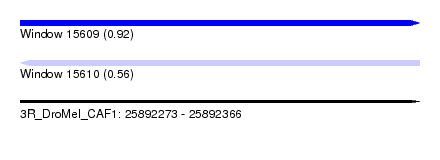
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 25,892,273 – 25,892,366 |
| Length | 93 |
| Max. P | 0.921717 |

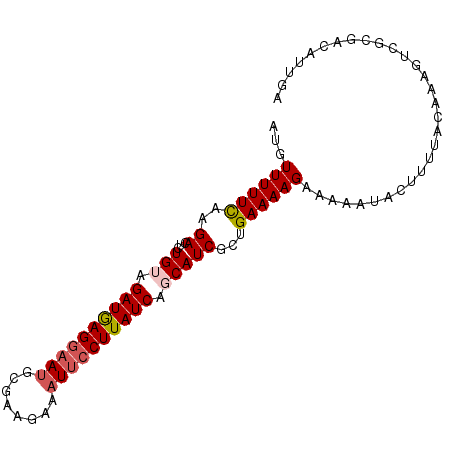
| Location | 25,892,273 – 25,892,366 |
|---|---|
| Length | 93 |
| Sequences | 5 |
| Columns | 93 |
| Reading direction | forward |
| Mean pairwise identity | 82.35 |
| Mean single sequence MFE | -20.74 |
| Consensus MFE | -14.28 |
| Energy contribution | -14.88 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.27 |
| Structure conservation index | 0.69 |
| SVM decision value | 1.14 |
| SVM RNA-class probability | 0.921717 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 25892273 93 + 27905053 AUGUUUUUUAAGACUCUGUAGAUGAGGAAUGCGUAGAAAUUCCUUAUCAGCAUCGUUGAAAAGAAAAAUACUUUUACAAAGUCGCGACAUUGA (((((......((((.((((((((((((((........))))))))))....((........))..........)))).))))..)))))... ( -21.80) >DroSec_CAF1 19372 91 + 1 AUGUUUUUCAAGAC--UGUAGAUGAGGAAUGCGAAGAAAUUCCUUAUCACCAUCGCUGAAAAGAAAAAUACUUUUACAAAGUCGCGACUUUGA ...(((((((.((.--((..((((((((((........))))))))))..))))..))))))).............((((((....)))))). ( -24.80) >DroSim_CAF1 19439 93 + 1 AUGUUUUUCAAGACUCUGUAGAUGAGGAAUGCGAAGAAAUUCCUUAUCACCAUCGCUGAAAAGAAAAAUACUUUUACAAAGUCGCGACAUUGA (((((......((((.((((((((((((((........)))))))))).......((....))...........)))).))))..)))))... ( -22.90) >DroEre_CAF1 27257 74 + 1 AUGUUUUUCAAGACUCUGUAGAUAAGGAAUGUGCAGAAAUUCCUUAUCAGCAUCGCCGAAAAGAAGAUUACUUU------------------- ...((((((..((...(((.((((((((((........)))))))))).)))))...))))))...........------------------- ( -18.00) >DroYak_CAF1 26675 92 + 1 AUGUUUUUCAAGAGUCUGUAGAUAAGAAAAGCAAAGAAAUGCCUUAUCAGCAUCGCCGAAAAGAAAAAUACUUUUACC-UGCCGCGACUUUGA ..........(((((((((.((((((....(((......))))))))).)))..((..(((((.......)))))...-.))...)))))).. ( -16.20) >consensus AUGUUUUUCAAGACUCUGUAGAUGAGGAAUGCGAAGAAAUUCCUUAUCAGCAUCGCUGAAAAGAAAAAUACUUUUACAAAGUCGCGACAUUGA ...((((((..((...(((.((((((((((........)))))))))).)))))...)))))).............................. (-14.28 = -14.88 + 0.60)

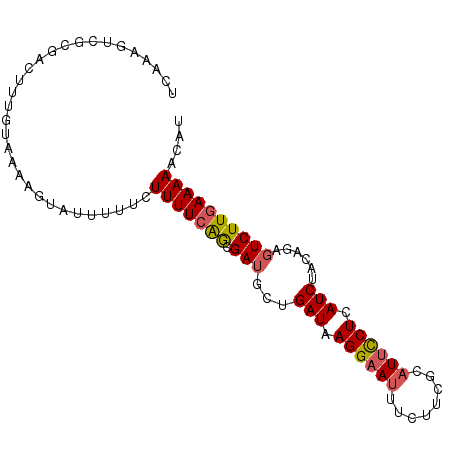
| Location | 25,892,273 – 25,892,366 |
|---|---|
| Length | 93 |
| Sequences | 5 |
| Columns | 93 |
| Reading direction | reverse |
| Mean pairwise identity | 82.35 |
| Mean single sequence MFE | -19.98 |
| Consensus MFE | -10.54 |
| Energy contribution | -10.78 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.10 |
| Structure conservation index | 0.53 |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.557024 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 25892273 93 - 27905053 UCAAUGUCGCGACUUUGUAAAAGUAUUUUUCUUUUCAACGAUGCUGAUAAGGAAUUUCUACGCAUUCCUCAUCUACAGAGUCUUAAAAAACAU ..........(((((((((..((((((.((......)).))))))(((.((((((........)))))).))))))))))))........... ( -21.20) >DroSec_CAF1 19372 91 - 1 UCAAAGUCGCGACUUUGUAAAAGUAUUUUUCUUUUCAGCGAUGGUGAUAAGGAAUUUCUUCGCAUUCCUCAUCUACA--GUCUUGAAAAACAU .((((((....))))))..............(((((((.(((.(((((.((((((........)))))).))).)).--)))))))))).... ( -20.30) >DroSim_CAF1 19439 93 - 1 UCAAUGUCGCGACUUUGUAAAAGUAUUUUUCUUUUCAGCGAUGGUGAUAAGGAAUUUCUUCGCAUUCCUCAUCUACAGAGUCUUGAAAAACAU ......(((.(((((((((((((.......))))...........(((.((((((........)))))).)))))))))))).)))....... ( -19.90) >DroEre_CAF1 27257 74 - 1 -------------------AAAGUAAUCUUCUUUUCGGCGAUGCUGAUAAGGAAUUUCUGCACAUUCCUUAUCUACAGAGUCUUGAAAAACAU -------------------............((((((((..((..((((((((((........))))))))))..))..)))..))))).... ( -18.70) >DroYak_CAF1 26675 92 - 1 UCAAAGUCGCGGCA-GGUAAAAGUAUUUUUCUUUUCGGCGAUGCUGAUAAGGCAUUUCUUUGCUUUUCUUAUCUACAGACUCUUGAAAAACAU ((((((((...(.(-(((((..........(((.(((((...))))).)))(((......))).....)))))).).)))).))))....... ( -19.80) >consensus UCAAAGUCGCGACUUUGUAAAAGUAUUUUUCUUUUCAGCGAUGCUGAUAAGGAAUUUCUUCGCAUUCCUCAUCUACAGAGUCUUGAAAAACAU ...............................(((((((.(((...(((.((((((........)))))).)))......)))))))))).... (-10.54 = -10.78 + 0.24)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:41:51 2006