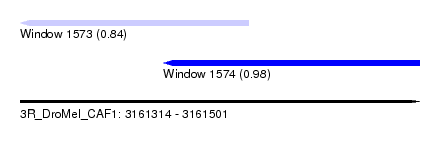
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 3,161,314 – 3,161,501 |
| Length | 187 |
| Max. P | 0.981716 |

| Location | 3,161,314 – 3,161,421 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 84.67 |
| Mean single sequence MFE | -33.04 |
| Consensus MFE | -23.15 |
| Energy contribution | -22.83 |
| Covariance contribution | -0.32 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.16 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.76 |
| SVM RNA-class probability | 0.843938 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 3161314 107 - 27905053 CCUGCAUUUAGGCGCUUCAGAAGUGAUUGCUACUAAACGAAGAAACCAGAAGACC-------GAAGACAGCCAACUUGCAACUGCAAA------GCUGCAAGCUGCAGGCGCCGAGAUGA ....(((((.((((((((((.((((.....)))).....................-------............((((((.((....)------).))))))))).))))))).))))). ( -29.00) >DroSec_CAF1 9189 106 - 1 CCUGCAUUUAGGCGCUUCGGCAGUGAUUGCUACCAACCGAAGA-ACCAGAAGACU-------GAAGACAGCCAACUCGCAACUGCAAA------GUUGCAAACUGCAGGCGCCGAGAUGA ....(((((.((((((...(((((...................-.......(.((-------(....))).).....((((((....)------)))))..))))).)))))).))))). ( -33.00) >DroSim_CAF1 11359 107 - 1 CCUGCAUUUAGGCGCUUCGGCAGUGAUUGCUACCAACCGAAGAAACCAGAAGACU-------GAAGACAGCCAACUCGCAACUGCAAA------GCUGCAAACUGCAGGCGCCGAGAUGA ....(((((.((((((...(((((....(((....................(.((-------(....))).).....((....))..)------)).....))))).)))))).))))). ( -29.80) >DroEre_CAF1 976 92 - 1 CCUGCAUUUAGGCGCUUCGGCAGUGAUUGCUACCAAGCAAAGAAACCAGAAG--------------ACUGCCAACUCGCAACUGCA--------------AACUGCAGGCGCCGAGAUGA ....(((((.(((((...((((((..(((((....))))).(....).....--------------)))))).........((((.--------------....))))))))).))))). ( -32.90) >DroYak_CAF1 10764 120 - 1 CCUGCAUUUAGGCGCUUUGGCAGUGAUUGCUACCAACCAAAGCAACCAGAAGACUGAAAACUGAAGACUGCCAACUCGCAACUGCAAACUGCAAACUGCAAACUGCAGGCGCCGAGAUGA ....(((((.(((((.((((((((..(((((.........))))).(((....)))..........)))))))).......(((((...(((.....)))...)))))))))).))))). ( -40.50) >consensus CCUGCAUUUAGGCGCUUCGGCAGUGAUUGCUACCAACCGAAGAAACCAGAAGACU_______GAAGACAGCCAACUCGCAACUGCAAA______GCUGCAAACUGCAGGCGCCGAGAUGA ....(((((.(((((...((.(((....))).))...................................((......))..(((((.................)))))))))).))))). (-23.15 = -22.83 + -0.32)

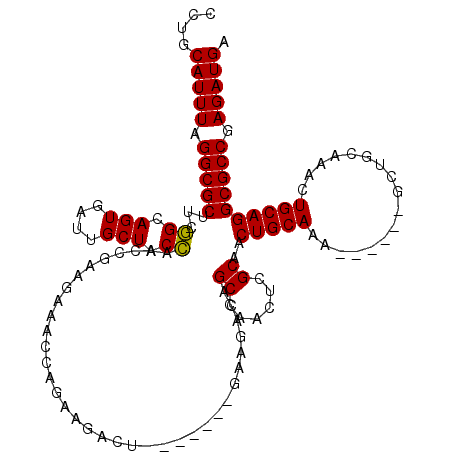
| Location | 3,161,381 – 3,161,501 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.75 |
| Mean single sequence MFE | -44.28 |
| Consensus MFE | -39.10 |
| Energy contribution | -39.62 |
| Covariance contribution | 0.52 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.88 |
| SVM decision value | 1.89 |
| SVM RNA-class probability | 0.981716 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 3161381 120 - 27905053 GAGGUGCUGGCUAGUAAUUAAGGCGACAAACAGUCAGAACGGGGCGUGGCCGGCCAUCUGACAUGUGCGAUGAUGUAUGGCCUGCAUUUAGGCGCUUCAGAAGUGAUUGCUACUAAACGA (((((((((((((...((((...((....(((((((((...((.((....)).)).)))))).))).)).))))...)))))........))))))))((.(((....))).))...... ( -37.20) >DroSec_CAF1 9255 120 - 1 GAGGUGCUGGCUAGUAAUUAAGGCGACAAACAGUCAGAACGGGGCGUGGCCAGCCAUCUGGCAUGUGCGAUGAUGUAUGUCCUGCAUUUAGGCGCUUCGGCAGUGAUUGCUACCAACCGA ..(((..(((.(((((((((..(((((.....)))....((((((((.(((((....)))))..(((((..(((....))).)))))....))))))))))..))))))))))))))).. ( -45.90) >DroSim_CAF1 11426 120 - 1 GAGGUGCUGGCUAGUAAUUAAGGCGACAAACAGUCAGAACGGGGCGUGGCCAGCCAUCUGGCAUGUGCGAUGAUGUAUGGCCUGCAUUUAGGCGCUUCGGCAGUGAUUGCUACCAACCGA ..(((..(((.(((((((((..(((((.....)))....((((((..(((((..(((((.((....)).).))))..))))).((......))))))))))..))))))))))))))).. ( -49.10) >DroEre_CAF1 1028 120 - 1 GAGGCGCUGGCUAGUAAUUAAGGCGACAAAGAGUCAGAACGGGGCGUGGUCGGCCAUCUGGCAUGUGCGAUGAUGUAUGGCCUGCAUUUAGGCGCUUCGGCAGUGAUUGCUACCAAGCAA .....(((((.(((((((((..(((((.....)))....((((((..(((((..(((((.((....)).).))))..))))).((......))))))))))..))))))))))).))).. ( -45.80) >DroYak_CAF1 10844 120 - 1 GAGGUGCUGGCUGGUAAUUAAGGCGACAAAGAGUCAGAACGGGGCGUGGUCGGCCAUCUGGCAUGUGCGAUGAUGUAUGGCCUGCAUUUAGGCGCUUUGGCAGUGAUUGCUACCAACCAA ..(((..(((.(((((((((..(((((.....)))....((((((..(((((..(((((.((....)).).))))..))))).((......))))))))))..))))))))))))))).. ( -43.40) >consensus GAGGUGCUGGCUAGUAAUUAAGGCGACAAACAGUCAGAACGGGGCGUGGCCGGCCAUCUGGCAUGUGCGAUGAUGUAUGGCCUGCAUUUAGGCGCUUCGGCAGUGAUUGCUACCAACCGA ..(((..(((.(((((((((..(((((.....)))....((((((..(((((..(((((.((....)).).))))..))))).((......))))))))))..))))))))))))))).. (-39.10 = -39.62 + 0.52)

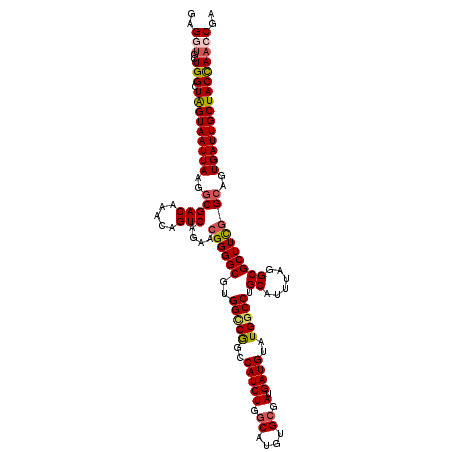
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:57:16 2006