
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 25,871,232 – 25,871,372 |
| Length | 140 |
| Max. P | 0.792620 |

| Location | 25,871,232 – 25,871,349 |
|---|---|
| Length | 117 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.45 |
| Mean single sequence MFE | -37.50 |
| Consensus MFE | -32.67 |
| Energy contribution | -32.40 |
| Covariance contribution | -0.27 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.68 |
| Structure conservation index | 0.87 |
| SVM decision value | 0.57 |
| SVM RNA-class probability | 0.784411 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 25871232 117 - 27905053 ---CAGCACAAAUGGCGCAAGCCCAAGGGUAUCGACAACAGAGUGCGUCGCCGCUUCAAGGGACAGUAUCUGAUGCCCAACAUCGGUUACGGAUCGAACAAGCGCACCCGCCACAUGCUG ---(((((.....(((....)))...((((.((.......))(((((((.((.......))))).(((.((((((.....)))))).)))...........))))))))......))))) ( -39.60) >DroVir_CAF1 28141 117 - 1 ---UAGCACAAAUGGCGCAAGCCCAAGGGUAUUGACAACAGAGUGCGUCGUCGCUUCAAGGGCCAAUAUCUGAUGCCCAACAUCGGUUACGGUUCCAACAAGCGCACCCGCCACAUGCUA ---(((((.....(((....)))...((((...(((.((...))..)))(.(((((...(((((.....((((((.....))))))....)))))....))))))))))......))))) ( -36.40) >DroPse_CAF1 13968 117 - 1 ---UAGCACAAAUGGCGCAAGCCUAAGGGUAUUGACAACAGAGUGCGUCGUCGCUUCAAGGGCCAAUACUUGAUGCCCAACAUCGGUUACGGCUCCAACAAGCGUACCCGUCAUAUGCUG ---(((((.....(((....)))...((((((.(((.((.......)).)))((((...(((((...(((.((((.....)))))))...)))))....))))))))))......))))) ( -39.30) >DroGri_CAF1 27101 117 - 1 ---CAGCACAAAUGGCGCAAGCCAAAGGGUAUCGACAACAGAGUACGUCGUCGCUUCAAGGGACAGUAUCUGAUGCCAAACAUUGGUUACGGCUCCAACCAGCGCACCCGCCACAUGCUG ---(((((....((((....))))..((((...(((..........)))(.((((....((..(.(((.(..(((.....)))..).))).)..))....)))))))))......))))) ( -38.20) >DroWil_CAF1 15730 120 - 1 GCUUAGCACAAAUGGCGCAAACCCAAGGGUAUUGACAACAGAGUACGUCGUCGCUUCAAGGGCCAAUAUCUGAUGCCAAACAUCGGUUAUGGUUCAAACAAGCGUACCCUCCACAUGCUG ...(((((....(((.(....))))(((((((.(((.((.......)).)))((((...((((((.((.((((((.....)))))).))))))))....))))))))))).....))))) ( -33.90) >DroPer_CAF1 14507 117 - 1 ---UAGCACAAAUGGCGCAAGCCUAAGGGUAUUGACAACAGAGUGCGUCGUCGCUUCAAGGGCCAAUACUUGAUGCCCAACAUCGGUUACGGCUCCAACAAGCGCACCCGUCAUAUGCUG ---(((((.....(((....)))...((((...(((.((...))..)))(.(((((...(((((...(((.((((.....)))))))...)))))....))))))))))......))))) ( -37.60) >consensus ___UAGCACAAAUGGCGCAAGCCCAAGGGUAUUGACAACAGAGUGCGUCGUCGCUUCAAGGGCCAAUAUCUGAUGCCCAACAUCGGUUACGGCUCCAACAAGCGCACCCGCCACAUGCUG ...(((((.....(((....)))...((((..((((..........)))).(((((...(((((.....((((((.....))))))....)))))....))))).))))......))))) (-32.67 = -32.40 + -0.27)

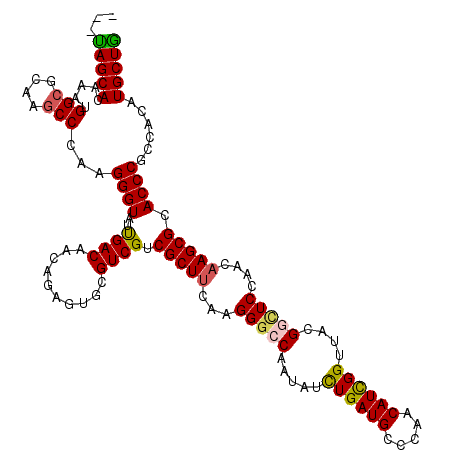
| Location | 25,871,272 – 25,871,372 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 79.36 |
| Mean single sequence MFE | -28.23 |
| Consensus MFE | -20.98 |
| Energy contribution | -20.90 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.43 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.59 |
| SVM RNA-class probability | 0.792620 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 25871272 100 - 27905053 UGU--UU----AAUCAACAU--G-UC-U-CCUUGCAGCACAAAUGGCGCAAGCCCAAGGGUAUCGACAACAGAGUGCGUCGCCGCUUCAAGGGACAGUAUCUGAUGCCCAA ...--..----.((((...(--(-((-.-(((((.(((......(((....)))....(((..((((......)).))..)))))).))))))))).....))))...... ( -31.70) >DroVir_CAF1 28181 106 - 1 AAC--AUUAUUAACCGGUUU--AUAUUC-CAUUUUAGCACAAAUGGCGCAAGCCCAAGGGUAUUGACAACAGAGUGCGUCGUCGCUUCAAGGGCCAAUAUCUGAUGCCCAA ...--..........((...--.....)-)..............(((....)))...((((((((((.((.......)).)))(((.....)))........))))))).. ( -23.10) >DroGri_CAF1 27141 105 - 1 AGA--A--AUUAAUCGGCUUUAAUUU-U-AAUUGCAGCACAAAUGGCGCAAGCCAAAGGGUAUCGACAACAGAGUACGUCGUCGCUUCAAGGGACAGUAUCUGAUGCCAAA .((--(--(((((......)))))))-)-..............((((....))))...(((((((((..........)))(((.(.....).))).......))))))... ( -27.60) >DroMoj_CAF1 32525 104 - 1 AAU--ACUAA-AAUUUACUC--AUAU-G-CAUUGCAGCACAAAUGGCGCAAGCCCAAGGGUAUCGACAACAGAGUACGUCGUCGCUUCAAGGGCCAAUAUCUGAUGCCCAA ...--.....-.........--...(-(-(......))).....(((....)))...((((((((((.((.......)).)))(((.....)))........))))))).. ( -26.50) >DroAna_CAF1 23434 102 - 1 UGCCACU----AAAGGAAUU--A-UC-U-GGUUGCAGCACAAAUGGCGCAAGCCCAAGGGUAUCGACAACAGAGUGCGUCGUCGCUUCAAGGGUCAGUACCUGAUGCCCAA (((.(((----...(((...--.-))-)-))).)))........(((....)))...(((((((.......(((.(((....))))))..((((....))))))))))).. ( -31.10) >DroPer_CAF1 14547 101 - 1 ACC--AU----AAUCAUGGU--C-UU-UAUGUUGUAGCACAAAUGGCGCAAGCCUAAGGGUAUUGACAACAGAGUGCGUCGUCGCUUCAAGGGCCAAUACUUGAUGCCCAA ...--..----.((((((((--(-((-(.((((((...((....(((....))).....))....))))))(((.(((....)))))))))))))).....))))...... ( -29.40) >consensus AGC__AU____AAUCGACUU__A_UU_U_CAUUGCAGCACAAAUGGCGCAAGCCCAAGGGUAUCGACAACAGAGUGCGUCGUCGCUUCAAGGGCCAAUAUCUGAUGCCCAA ............................................(((....)))...(((((((.......(((.(((....)))))).(((.......)))))))))).. (-20.98 = -20.90 + -0.08)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:41:46 2006