
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 25,867,055 – 25,867,197 |
| Length | 142 |
| Max. P | 0.773076 |

| Location | 25,867,055 – 25,867,157 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 65.37 |
| Mean single sequence MFE | -37.43 |
| Consensus MFE | -11.52 |
| Energy contribution | -11.67 |
| Covariance contribution | 0.14 |
| Combinations/Pair | 1.56 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.31 |
| SVM decision value | 0.29 |
| SVM RNA-class probability | 0.669101 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
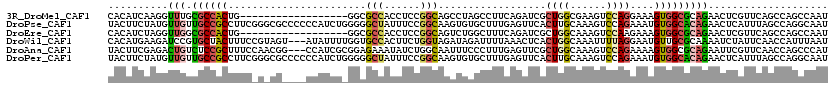
>3R_DroMel_CAF1 25867055 102 - 27905053 CACAUCAAGGUUUGCGCCACUG------------------GGCGCCACCUCCGGCAGCCUAGCCUUCAGAUCGCUGGCGAAGUCCAGGAAAGUGGCGCAGAACUCGUUCAGCCAGCCAAU ........((((((((((((((------------------((((((......))).(((.(((.........))))))...)))).....)))))))))(((....)))))))....... ( -38.00) >DroPse_CAF1 10157 120 - 1 UACUUCUAUGUUGUUGCCGCCUUCGGGCGCCCCCCAUCUGGGGGCUAUUUCCGGCAAGUGUGCUUUGAGUUCACUUGCAAAGUCCAGAAAUGUGGCACAGAACUCAUUUAGCCAGGCAAU ........((((.((((((((....)))((((((.....)))))).......))))).((.(((.(((((((...(((......((....))..)))..)))))))...))))))))).. ( -46.30) >DroEre_CAF1 19206 102 - 1 CACAUCUAGGUUGGCGCCACUG------------------GGCGCCACCUCCGGCAGUCUGGCUUUCAGAUCGCUGGCAAAGUCCAAGAAAGUGGCGCAGAACUCGUUCAGCCAGCCAAU ........((((((((((((((------------------(((.......(((((.(((((.....))))).)))))....)))).....))))))...(((....))).)))))))... ( -43.10) >DroWil_CAF1 11653 117 - 1 CACAUGAAGAUCCGUGCUACUUUCCGUAGU---AUAUUUUGGUGCCACUUCUGGUAGAUAGAUUUUAAACUCACUGGCAAAUUUUAGGAAUGUUGCGCAAAAUCUAUUCAACCAUUUAAU ..(((.(((((..(((((((.....)))))---))))))).))).......((((.((((((((((.(((...((((......))))....)))....))))))))))..))))...... ( -24.40) >DroAna_CAF1 19371 117 - 1 UACUUCGAGACUGUCUCCGCUUUCCAACGG---CCAUCGCGGAGAAAUAUCUGGCAAUUUCCCUUUGAGUUCGCUGGCAAAGUCCAGAAAAGUGGCGCAGAAUUCGUUCAACCAGCCCAU ...(((..((((.(((((((..........---.....))))))).....(..(((((((......))))).))..)...))))..)))..(.(((...(((....))).....)))).. ( -26.46) >DroPer_CAF1 10571 120 - 1 UACUUCUAUGUUGUUGCCGCCUUCGGGCGCCCCCCAUCUGGGGGCUAUUUCCGGCAAGUGUGCUUUGAGUUCACUUGCAAAGUCCAGAAAUGUGGCACAGAACUCAUUUAGCCAGGCAAU ........((((.((((((((....)))((((((.....)))))).......))))).((.(((.(((((((...(((......((....))..)))..)))))))...))))))))).. ( -46.30) >consensus CACAUCUAGGUUGGCGCCACUUUC_G_CG____CCAUCU_GGCGCCACUUCCGGCAAGUUGGCUUUGAGUUCACUGGCAAAGUCCAGAAAAGUGGCGCAGAACUCAUUCAGCCAGCCAAU ..........(((.((((((.......................(((......)))..................((((......))))....))))))))).................... (-11.52 = -11.67 + 0.14)


| Location | 25,867,095 – 25,867,197 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 67.74 |
| Mean single sequence MFE | -41.64 |
| Consensus MFE | -12.62 |
| Energy contribution | -11.18 |
| Covariance contribution | -1.44 |
| Combinations/Pair | 1.62 |
| Mean z-score | -2.06 |
| Structure conservation index | 0.30 |
| SVM decision value | 0.53 |
| SVM RNA-class probability | 0.773076 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
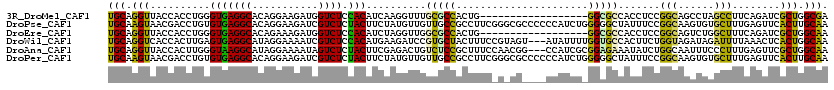
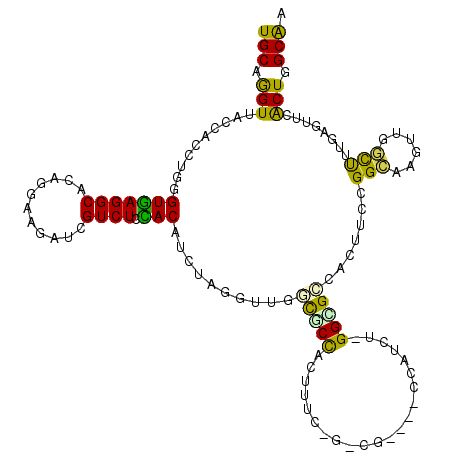
>3R_DroMel_CAF1 25867095 102 - 27905053 UGCAGGUUACCACCUGGGUGAGGCACAGGAAGAUGGUCUCCACAUCAAGGUUUGCGCCACUG------------------GGCGCCACCUCCGGCAGCCUAGCCUUCAGAUCGCUGGCGA ..(((((....)))))(((.((((...(((.((((.......))))..(((..(((((....------------------))))).))))))....)))).))).((((....))))... ( -37.50) >DroPse_CAF1 10197 120 - 1 UGCAAGUAACGACCUGUGUGAGGCACAGGAAGAUCGUCUCUACUUCUAUGUUGUUGCCGCCUUCGGGCGCCCCCCAUCUGGGGGCUAUUUCCGGCAAGUGUGCUUUGAGUUCACUUGCAA ((((((((((........((((((((((((((..........)))))......((((((((....)))((((((.....)))))).......))))).))))))))).))).))))))). ( -50.60) >DroEre_CAF1 19246 102 - 1 UGCAGGUUACCACCUGGGUGAGGCACAGAAAGAUGGUCUCCACAUCUAGGUUGGCGCCACUG------------------GGCGCCACCUCCGGCAGUCUGGCUUUCAGAUCGCUGGCAA (((((((....(((((((((((((.((......)))))))...)))))))).((((((....------------------)))))))))).((((.(((((.....))))).))))))). ( -45.60) >DroWil_CAF1 11693 117 - 1 UGCAGGUCACCACUUGAGUGAGGCAUAGGAAAAUCGUCUCCACAUGAAGAUCCGUGCUACUUUCCGUAGU---AUAUUUUGGUGCCACUUCUGGUAGAUAGAUUUUAAACUCACUGGCAA ..(((((....)))))((((((.......((((((((((((((((.(((((..(((((((.....)))))---))))))).))).......))).)))).))))))...))))))..... ( -30.21) >DroAna_CAF1 19411 117 - 1 UGCAGGUUACCACUUGGGUAAGGCAUAGGAAAAUAGUCUCUACUUCGAGACUGUCUCCGCUUUCCAACGG---CCAUCGCGGAGAAAUAUCUGGCAAUUUCCCUUUGAGUUCGCUGGCAA (((.(((.((((...(((....((...(((...(((((((......)))))))(((((((..........---.....)))))))....))).)).....)))..)).))..))).))). ( -35.36) >DroPer_CAF1 10611 120 - 1 UGCAAGUAACGACCUGUGUGAGGCACAGGAAGAUCGUCUCUACUUCUAUGUUGUUGCCGCCUUCGGGCGCCCCCCAUCUGGGGGCUAUUUCCGGCAAGUGUGCUUUGAGUUCACUUGCAA ((((((((((........((((((((((((((..........)))))......((((((((....)))((((((.....)))))).......))))).))))))))).))).))))))). ( -50.60) >consensus UGCAGGUUACCACCUGGGUGAGGCACAGGAAGAUCGUCUCCACAUCUAGGUUGGCGCCACUUUC_G_CG____CCAUCU_GGCGCCACUUCCGGCAAGUUGGCUUUGAGUUCACUGGCAA (((.(((..........(((((((...........)))).)))..........(((((......................))))).......(((......)))........))).))). (-12.62 = -11.18 + -1.44)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:41:43 2006