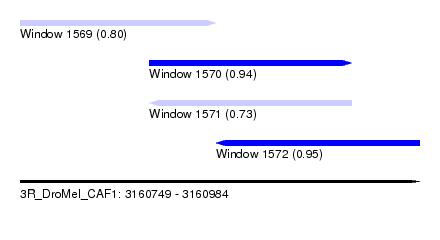
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 3,160,749 – 3,160,984 |
| Length | 235 |
| Max. P | 0.946461 |

| Location | 3,160,749 – 3,160,864 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.79 |
| Mean single sequence MFE | -43.00 |
| Consensus MFE | -37.68 |
| Energy contribution | -37.88 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.36 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.60 |
| SVM RNA-class probability | 0.795141 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 3160749 115 + 27905053 UGUCAGCCCUUACGGGGCCAGGAUUAUGUUAAACAGUUCGCCAGGGCCCCAGGAACCCGUCC----GACUGCCAGUCUGUCUG-CGAGACCCUCGACUCCAGUCUUUGGGGGUCUCAUUG .(((.........((((((.((....((.....)).....))..)))))).(((.....)))----))).((.((.....)))-)((((((((((((....)))...))))))))).... ( -43.00) >DroSec_CAF1 8592 116 + 1 UGUCAGCCCUUACGGGGCCAGGAUUAUGUUAAACAGUACGCCAGGGCCCCAGGAACCCGUCC----GACCGCCAGUCUGUCUGUCGAGACCCUCGACUCCAGUCUUUGGGGGUCUCAUUG .............((((((.((....((.....)).....))..)))))).(((.....)))----((((.((((.(((...((((((...))))))..)))...)))).))))...... ( -43.10) >DroSim_CAF1 10802 116 + 1 UGUCAGCCCUUACGGGGCCAGGAUUAUGUUAAACAGUUCGCCAGGGCCCCAGGAACCCGUCC----GACGGCCAGUCUGUCUGUCGAGACCCUCGACUCCAGUCUUUGGGGGUCUCAUUG .(((.........((((((.((....((.....)).....))..)))))).(((.....)))----)))(((.((.....)))))((((((((((((....)))...))))))))).... ( -44.40) >DroEre_CAF1 444 113 + 1 UGUCAGCCCUUACGGGGCCAGGAUUAUGUUAAACAGUUCGCA-GGGCCCCAAGAACCCAG------GACUGCCAGUCUGUCUGUCGAGACCCUCGACUCCAGUCUUUGGGGGUCUCAUUG .(.(((.......((((((.(((((.(.....).)))))...-.)))))).........(------(((.....))))..))).)((((((((((((....)))...))))))))).... ( -41.00) >DroYak_CAF1 10171 119 + 1 UGUCAGCCCUUACGGGGCCAGGAUUAUGUUAAACAGUUCGCC-AGGCCCCAAGAACCCGGCCAACUGACUGCCAGUCUGUCUGUCGAGACCCUCGACUUCAGUCUUUGGGGGUCUCAUUG .............((((((.((....((.....)).....))-.))))))........(((..((.(((.....))).))..)))((((((((((((....)))...))))))))).... ( -43.50) >consensus UGUCAGCCCUUACGGGGCCAGGAUUAUGUUAAACAGUUCGCCAGGGCCCCAGGAACCCGUCC____GACUGCCAGUCUGUCUGUCGAGACCCUCGACUCCAGUCUUUGGGGGUCUCAUUG .(.(((.......((((((.((....((.....)).....))..))))))................(((.....)))...))).)((((((((((((....)))...))))))))).... (-37.68 = -37.88 + 0.20)

| Location | 3,160,825 – 3,160,944 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.50 |
| Mean single sequence MFE | -39.04 |
| Consensus MFE | -37.16 |
| Energy contribution | -37.36 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.48 |
| Structure conservation index | 0.95 |
| SVM decision value | 1.30 |
| SVM RNA-class probability | 0.939133 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
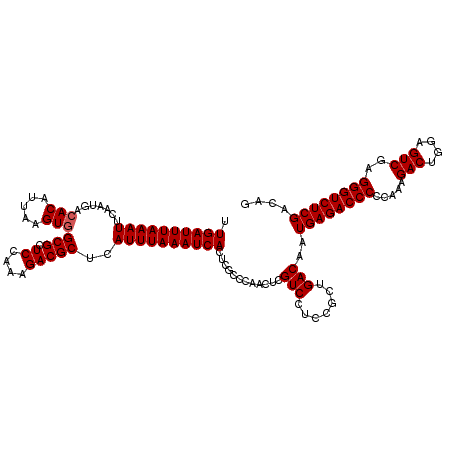
>3R_DroMel_CAF1 3160825 119 + 27905053 CUG-CGAGACCCUCGACUCCAGUCUUUGGGGGUCUCAUUGUCAGCGGAGGACGAGUUGGGCGAGUGAUUUAAAUGAGCGUCUUUAGAGCGCCACUUAAUGUGUCAUUGAAUUUAAAUCAA .((-(((((((((((((....)))...))))))))).(((((.......))))).....)))..((((((((((..(((((....)).)))(((.....))).......)))))))))). ( -37.80) >DroSec_CAF1 8668 120 + 1 CUGUCGAGACCCUCGACUCCAGUCUUUGGGGGUCUCAUUGUCAGCGGAGGACGAGUUGGGCGAGUGAUUUAAAUGAGCGUCUUUGGAACGCCACUUAAUGUGUCAUUGAAUUUAAAUCAA .((((((((((((((((....)))...))))))))).(((((.......)))))....))))..((((((((((..(((((....).))))(((.....))).......)))))))))). ( -38.50) >DroSim_CAF1 10878 120 + 1 CUGUCGAGACCCUCGACUCCAGUCUUUGGGGGUCUCAUUGUCAGCGGAGGACGAGUUGGGCGAGUGAUUUAAAUGAGCGUCUUUGGAGCGCCACUUAAUGUCUCAUUGAAUUUAAAUCAA .((((((((((((((((....)))...))))))))).(((((.......)))))....))))..(((((((((((.(((((....)).))))..((((((...)))))))))))))))). ( -38.20) >DroEre_CAF1 517 120 + 1 CUGUCGAGACCCUCGACUCCAGUCUUUGGGGGUCUCAUUGUCAGCGGAGGACGAGUUGGGCGAGUGAUUUAAAUGAGCGUCUCUGGAGCGCCACUUAAUGUGUCAUUGAAUUUAAAUCAA .((((((((((((((((....)))...))))))))).(((((.......)))))....))))..((((((((((..(((.((....)))))(((.....))).......)))))))))). ( -38.80) >DroYak_CAF1 10250 120 + 1 CUGUCGAGACCCUCGACUUCAGUCUUUGGGGGUCUCAUUGUCAGCGGAGGACGAGUUGGGCGAGUGAUUUAAAUGGGCGUCUUUAGAGCGCCACUUAAUGUGUCAUUGAAUUUAAAUCAA .((((((((((((((((....)))...))))))))).(((((.......)))))....))))..((((((((((.((((((....)).))))..((((((...)))))))))))))))). ( -41.90) >consensus CUGUCGAGACCCUCGACUCCAGUCUUUGGGGGUCUCAUUGUCAGCGGAGGACGAGUUGGGCGAGUGAUUUAAAUGAGCGUCUUUGGAGCGCCACUUAAUGUGUCAUUGAAUUUAAAUCAA .((((((((((((((((....)))...))))))))).(((((.......)))))....))))..(((((((((((.(((((....)).))))..((((((...)))))))))))))))). (-37.16 = -37.36 + 0.20)

| Location | 3,160,825 – 3,160,944 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.50 |
| Mean single sequence MFE | -30.88 |
| Consensus MFE | -28.56 |
| Energy contribution | -28.76 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.30 |
| Structure conservation index | 0.92 |
| SVM decision value | 0.42 |
| SVM RNA-class probability | 0.730569 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 3160825 119 - 27905053 UUGAUUUAAAUUCAAUGACACAUUAAGUGGCGCUCUAAAGACGCUCAUUUAAAUCACUCGCCCAACUCGUCCUCCGCUGACAAUGAGACCCCCAAAGACUGGAGUCGAGGGUCUCG-CAG .((((((((((.......(((.....)))(((.((....)))))..))))))))))...((.......(((.......)))...(((((((.....(((....)))..))))))))-).. ( -31.90) >DroSec_CAF1 8668 120 - 1 UUGAUUUAAAUUCAAUGACACAUUAAGUGGCGUUCCAAAGACGCUCAUUUAAAUCACUCGCCCAACUCGUCCUCCGCUGACAAUGAGACCCCCAAAGACUGGAGUCGAGGGUCUCGACAG .((((((((((.......(((.....)))(((((.....)))))..))))))))))............(((.......)))..((((((((.....(((....)))..)))))))).... ( -31.50) >DroSim_CAF1 10878 120 - 1 UUGAUUUAAAUUCAAUGAGACAUUAAGUGGCGCUCCAAAGACGCUCAUUUAAAUCACUCGCCCAACUCGUCCUCCGCUGACAAUGAGACCCCCAAAGACUGGAGUCGAGGGUCUCGACAG ((((.......)))).(((...((((((((((((....)).))).)))))))....))).........(((.......)))..((((((((.....(((....)))..)))))))).... ( -29.60) >DroEre_CAF1 517 120 - 1 UUGAUUUAAAUUCAAUGACACAUUAAGUGGCGCUCCAGAGACGCUCAUUUAAAUCACUCGCCCAACUCGUCCUCCGCUGACAAUGAGACCCCCAAAGACUGGAGUCGAGGGUCUCGACAG .((((((((((.......(((.....)))(((((....)).)))..))))))))))............(((.......)))..((((((((.....(((....)))..)))))))).... ( -30.70) >DroYak_CAF1 10250 120 - 1 UUGAUUUAAAUUCAAUGACACAUUAAGUGGCGCUCUAAAGACGCCCAUUUAAAUCACUCGCCCAACUCGUCCUCCGCUGACAAUGAGACCCCCAAAGACUGAAGUCGAGGGUCUCGACAG .((((((((((..((((...))))..(.((((.((....)))))))))))))))))............(((.......)))..((((((((.....(((....)))..)))))))).... ( -30.70) >consensus UUGAUUUAAAUUCAAUGACACAUUAAGUGGCGCUCCAAAGACGCUCAUUUAAAUCACUCGCCCAACUCGUCCUCCGCUGACAAUGAGACCCCCAAAGACUGGAGUCGAGGGUCUCGACAG .((((((((((.......(((.....)))(((.((....)))))..))))))))))............(((.......)))..((((((((.....(((....)))..)))))))).... (-28.56 = -28.76 + 0.20)

| Location | 3,160,864 – 3,160,984 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.83 |
| Mean single sequence MFE | -23.68 |
| Consensus MFE | -20.00 |
| Energy contribution | -20.24 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.78 |
| Structure conservation index | 0.84 |
| SVM decision value | 1.36 |
| SVM RNA-class probability | 0.946461 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
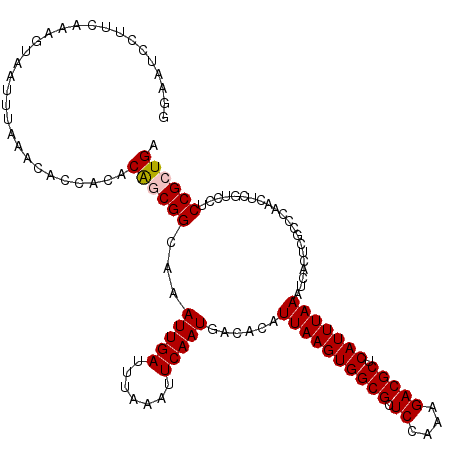
>3R_DroMel_CAF1 3160864 120 - 27905053 GGAAUCCUUCAAAGUAAUUUAAACACCACACAGCGGCAAAUUGAUUUAAAUUCAAUGACACAUUAAGUGGCGCUCUAAAGACGCUCAUUUAAAUCACUCGCCCAACUCGUCCUCCGCUGA ..............................((((((...(((((.......)))))......((((((((((.((....))))).))))))).....................)))))). ( -23.00) >DroSec_CAF1 8708 120 - 1 GAAAUCCUUCAAAGUAAUUUAAACACCACACAGCGGCAAAUUGAUUUAAAUUCAAUGACACAUUAAGUGGCGUUCCAAAGACGCUCAUUUAAAUCACUCGCCCAACUCGUCCUCCGCUGA ..............................((((((...(((((.......)))))......((((((((((((.....))))).))))))).....................)))))). ( -24.20) >DroSim_CAF1 10918 120 - 1 GGAAUCCUUCAAAGCAAUUUAAACACCACACAGCGGCAAAUUGAUUUAAAUUCAAUGAGACAUUAAGUGGCGCUCCAAAGACGCUCAUUUAAAUCACUCGCCCAACUCGUCCUCCGCUGA ..............................((((((...(((((.......)))))(((...((((((((((((....)).))).)))))))....)))..............)))))). ( -24.60) >DroEre_CAF1 557 120 - 1 GGAGUCCUUCAAAGUAAUUGAAACACCACACAACGGCAAAUUGAUUUAAAUUCAAUGACACAUUAAGUGGCGCUCCAGAGACGCUCAUUUAAAUCACUCGCCCAACUCGUCCUCCGCUGA ((((..((((((.....)))))............(((..(((((.......)))))......((((((((((((....)).))).))))))).......)))......)..))))..... ( -26.60) >DroYak_CAF1 10290 113 - 1 -------UUCAAAGUAAUUUAAACACCACACGACGGCAAAUUGAUUUAAAUUCAAUGACACAUUAAGUGGCGCUCUAAAGACGCCCAUUUAAAUCACUCGCCCAACUCGUCCUCCGCUGA -------......................((((.(((..(((((.......)))))......((((((((.((((....)).)))))))))).......)))....)))).......... ( -20.00) >consensus GGAAUCCUUCAAAGUAAUUUAAACACCACACAGCGGCAAAUUGAUUUAAAUUCAAUGACACAUUAAGUGGCGCUCCAAAGACGCUCAUUUAAAUCACUCGCCCAACUCGUCCUCCGCUGA ..............................((((((...(((((.......)))))......((((((((((.((....))))).))))))).....................)))))). (-20.00 = -20.24 + 0.24)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:57:14 2006