
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 25,847,471 – 25,847,814 |
| Length | 343 |
| Max. P | 0.993196 |

| Location | 25,847,471 – 25,847,589 |
|---|---|
| Length | 118 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.76 |
| Mean single sequence MFE | -41.26 |
| Consensus MFE | -35.44 |
| Energy contribution | -35.25 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.78 |
| SVM RNA-class probability | 0.848649 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 25847471 118 - 27905053 AGUGACGGAUUCGAUUCCCCGGGGAAUCCU-ACCAAUUAGUGCAAAUUCCGA-AAUGCACGGAAAAGCGAACGAAAGCUGCGAAUUCGUUACACUGCUGCACUUGGACUUUUCGGGGCGC .(((((((((((((((((....))))))..-..........((...(((((.-......))))).(((........))))))))))))))))...((.((.((((((...)))))))))) ( -39.20) >DroSec_CAF1 1914 118 - 1 AGUGACGGACUCGAUUCCCCGGGGAAUCCC-CCCAAUUUGUGCAUAAUCCGA-AAUGCGCGGAAAAGCGAACGAAAGCUGCGAAUUCGUUACACUGCUGCACUUGGACUUUUCGGGGCGC .((((((((.(((.((((..((((.....)-))).....((((((.......-.)))))))))).(((........))).))).))))))))...((.((.((((((...)))))))))) ( -40.30) >DroSim_CAF1 1896 119 - 1 AGUGACGGAUUCGAUUCCCCGGGGAAUCCCCCCCAAUUAGUGCAUAAUUCGA-AAUGCGCGGAAAAGCGAACGAAAGCUGCGAAUUCGUUACACUGCUGCACUUGGACUUUUCGGGGCGC .((((((((((((.((((..((((......)))).....((((((.......-.)))))))))).(((........))).))))))))))))...((.((.((((((...)))))))))) ( -44.40) >DroYak_CAF1 100 116 - 1 AGUGACGAAUUCGAUUCCCCGGGGAAUCCC-CCCAAGUGGUGUAUAUUCCGAAAAUGCGCGGUAAAGCGGACGAAAGCUGCGAAUUCGUUAC---GCUGCAUUUGAACUUUUGGGGGCGC .(((((((((((..((..((((((.....)-))).....((((((.........))))))))..))((((.(....))))))))))))))))---(((.(.............).))).. ( -41.12) >consensus AGUGACGGAUUCGAUUCCCCGGGGAAUCCC_CCCAAUUAGUGCAUAAUCCGA_AAUGCGCGGAAAAGCGAACGAAAGCUGCGAAUUCGUUACACUGCUGCACUUGGACUUUUCGGGGCGC .(((((((((((((((((....))))))...........((((((.........))))))......(((..(....).))))))))))))))...((.((.((((((...)))))))))) (-35.44 = -35.25 + -0.19)



| Location | 25,847,589 – 25,847,694 |
|---|---|
| Length | 105 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.33 |
| Mean single sequence MFE | -28.75 |
| Consensus MFE | -20.49 |
| Energy contribution | -23.55 |
| Covariance contribution | 3.06 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.16 |
| SVM RNA-class probability | 0.611995 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 25847589 105 + 27905053 UUUAAUCCUGUUUGCUGUUCUUUUCGCACU------------CGCUUUUGUUUUUGCAAUGCAG---GCUCCUGCACUUGCGGUACACACACAUACCAGCAGCUGCUACGGCCGCCGCUG ........(((.((((((.......(((..------------.((....))...)))..(((((---....)))))...)))))).))).......((((.((.((....)).)).)))) ( -30.30) >DroSec_CAF1 2032 105 + 1 UUUAGUCCUGUUUGCUGUUCUUUUCGCACU------------CGCUUUUGUUUUUGCAAUGCAG---GCUCCUGCACUUGCGGUACACACACAUACCAGCAGCUGCUACGGCCGCCGCUG ........(((.((((((.......(((..------------.((....))...)))..(((((---....)))))...)))))).))).......((((.((.((....)).)).)))) ( -30.30) >DroSim_CAF1 2015 105 + 1 UUUAGUCCUGUUUGCUGUUCUUUUCGCACU------------CGCUUUUGUUUUUGCAAUGCAG---GCUCCUGCACUUGCGGUACACACACAUACCAGCAGCUGCUACGGCCGCCGCUG ........(((.((((((.......(((..------------.((....))...)))..(((((---....)))))...)))))).))).......((((.((.((....)).)).)))) ( -30.30) >DroYak_CAF1 216 120 + 1 UUUAGUCCUGUUUCUUGUUUGUUUCGCACUUUACGCACUUUUCACUUUUGUUUUUGCAAUGCAAAAUGCUCCUGCACUUGCGAUACACACACAUACCAACAGCUGCUACGGCCGCCGCUG ..((((.(((((...(((.(((.(((((.......................((((((...))))))(((....)))..)))))...))).)))....)))))..))))((((....)))) ( -24.10) >consensus UUUAGUCCUGUUUGCUGUUCUUUUCGCACU____________CGCUUUUGUUUUUGCAAUGCAG___GCUCCUGCACUUGCGGUACACACACAUACCAGCAGCUGCUACGGCCGCCGCUG ........(((.((.(((.....(((((...................((((....))))(((((.......)))))..))))).))))).)))...((((.((.((....)).)).)))) (-20.49 = -23.55 + 3.06)

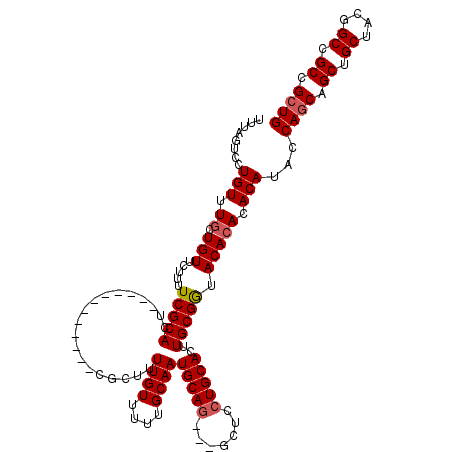

| Location | 25,847,589 – 25,847,694 |
|---|---|
| Length | 105 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.33 |
| Mean single sequence MFE | -33.85 |
| Consensus MFE | -27.62 |
| Energy contribution | -27.12 |
| Covariance contribution | -0.50 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.43 |
| Structure conservation index | 0.82 |
| SVM decision value | 1.49 |
| SVM RNA-class probability | 0.958124 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 25847589 105 - 27905053 CAGCGGCGGCCGUAGCAGCUGCUGGUAUGUGUGUGUACCGCAAGUGCAGGAGC---CUGCAUUGCAAAAACAAAAGCG------------AGUGCGAAAAGAACAGCAAACAGGAUUAAA (((((((.((....)).)))))))((.(((((.((((((((.(((((((....---)))))))(......)....)))------------.)))))......))).)).))......... ( -33.90) >DroSec_CAF1 2032 105 - 1 CAGCGGCGGCCGUAGCAGCUGCUGGUAUGUGUGUGUACCGCAAGUGCAGGAGC---CUGCAUUGCAAAAACAAAAGCG------------AGUGCGAAAAGAACAGCAAACAGGACUAAA (((((((.((....)).)))))))((.(((((.((((((((.(((((((....---)))))))(......)....)))------------.)))))......))).)).))......... ( -33.90) >DroSim_CAF1 2015 105 - 1 CAGCGGCGGCCGUAGCAGCUGCUGGUAUGUGUGUGUACCGCAAGUGCAGGAGC---CUGCAUUGCAAAAACAAAAGCG------------AGUGCGAAAAGAACAGCAAACAGGACUAAA (((((((.((....)).)))))))((.(((((.((((((((.(((((((....---)))))))(......)....)))------------.)))))......))).)).))......... ( -33.90) >DroYak_CAF1 216 120 - 1 CAGCGGCGGCCGUAGCAGCUGUUGGUAUGUGUGUGUAUCGCAAGUGCAGGAGCAUUUUGCAUUGCAAAAACAAAAGUGAAAAGUGCGUAAAGUGCGAAACAAACAAGAAACAGGACUAAA (((((((.((....)).)))))))((...(((.(((.(((((..(((....(((((((.((((...........)))).))))))))))...))))).))).)))....))......... ( -33.70) >consensus CAGCGGCGGCCGUAGCAGCUGCUGGUAUGUGUGUGUACCGCAAGUGCAGGAGC___CUGCAUUGCAAAAACAAAAGCG____________AGUGCGAAAAGAACAGCAAACAGGACUAAA (((((((.((....)).)))))))((.(((.(((....(((((((((((.......)))))))((..........))...............))))......)))))).))......... (-27.62 = -27.12 + -0.50)

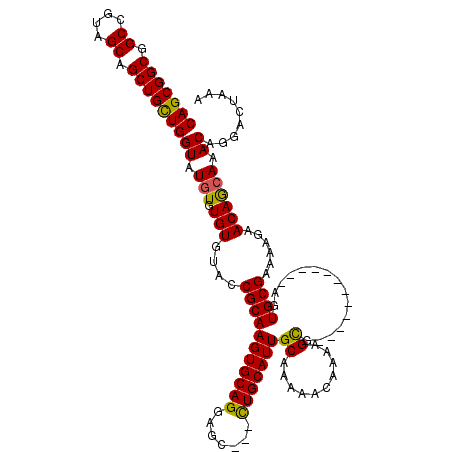

| Location | 25,847,654 – 25,847,774 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.00 |
| Mean single sequence MFE | -55.21 |
| Consensus MFE | -52.75 |
| Energy contribution | -53.19 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.87 |
| Structure conservation index | 0.96 |
| SVM decision value | 2.38 |
| SVM RNA-class probability | 0.993196 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 25847654 120 - 27905053 AAUGCGCGCGGCAGUGGGUUGAAGCGAGACGGGUAGAGCUAGAGAGAUGCCCGUUUGCCCGCUGUGCGUGUGUGUGUGUGCAGCGGCGGCCGUAGCAGCUGCUGGUAUGUGUGUGUACCG .((((((((((((((((((.......(((((((((...((....)).)))))))))))))))))).)))))))))....(((((.((.......)).))))).(((((......))))). ( -55.31) >DroSec_CAF1 2097 120 - 1 AAUGCGCGCGGCAGUGGGUUGAAGCGAGACGGGUAGAGCUAGAGCGAUGCCCGUUUGCCCGCUGUGCGUGUGUGUGUGUGCAGCGGCGGCCGUAGCAGCUGCUGGUAUGUGUGUGUACCG .((((((((((((((((((.......(((((((((..((....))..)))))))))))))))))).)))))))))....(((((.((.......)).))))).(((((......))))). ( -59.11) >DroSim_CAF1 2080 120 - 1 AAUGCGCGCGGCAGUGGGUUGAAGCGAGACGGGUAGAGCUAGAGCGAUGCCCGUUUGCCCGCUGUGCGUGUGUGUGUGUGCAGCGGCGGCCGUAGCAGCUGCUGGUAUGUGUGUGUACCG .((((((((((((((((((.......(((((((((..((....))..)))))))))))))))))).)))))))))....(((((.((.......)).))))).(((((......))))). ( -59.11) >DroYak_CAF1 296 114 - 1 AAUGCGCACGGCAGUGGGUUGAAGCGAGACGGGAAUA------GCGAUGCCCGUUUGCCCGCUGUGCGUGUGUGUGUGUGCAGCGGCGGCCGUAGCAGCUGUUGGUAUGUGUGUGUAUCG .((((((((((((((((((.......(((((((.((.------...)).)))))))))))))))).)))))))))(..(((((((((.((....)).)))))).)))..).......... ( -47.31) >consensus AAUGCGCGCGGCAGUGGGUUGAAGCGAGACGGGUAGAGCUAGAGCGAUGCCCGUUUGCCCGCUGUGCGUGUGUGUGUGUGCAGCGGCGGCCGUAGCAGCUGCUGGUAUGUGUGUGUACCG .((((((((((((((((((.......(((((((((..((....))..)))))))))))))))))).)))))))))....(((((.((.......)).))))).(((((......))))). (-52.75 = -53.19 + 0.44)



| Location | 25,847,694 – 25,847,814 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.92 |
| Mean single sequence MFE | -43.04 |
| Consensus MFE | -38.77 |
| Energy contribution | -39.59 |
| Covariance contribution | 0.81 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.89 |
| Structure conservation index | 0.90 |
| SVM decision value | 2.28 |
| SVM RNA-class probability | 0.991593 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 25847694 120 - 27905053 AUAAAUCAAGCGAAGUAGCAACAAAUGCGGGAAAUGUGAAAAUGCGCGCGGCAGUGGGUUGAAGCGAGACGGGUAGAGCUAGAGAGAUGCCCGUUUGCCCGCUGUGCGUGUGUGUGUGUG .....(((.........(((.....)))........)))..((((((((((((((((((.......(((((((((...((....)).)))))))))))))))))).)))))))))..... ( -41.84) >DroSec_CAF1 2137 120 - 1 AUAAAUCCAGCGAAGUAGCAACAAAAGCGGGAAAUGUGAAAAUGCGCGCGGCAGUGGGUUGAAGCGAGACGGGUAGAGCUAGAGCGAUGCCCGUUUGCCCGCUGUGCGUGUGUGUGUGUG .....(((.((...((....))....)).))).........((((((((((((((((((.......(((((((((..((....))..)))))))))))))))))).)))))))))..... ( -47.21) >DroSim_CAF1 2120 120 - 1 AUAAAUCAAGCGAAGUAGCAACAAAAGCGGGAAAUGUGAAAAUGCGCGCGGCAGUGGGUUGAAGCGAGACGGGUAGAGCUAGAGCGAUGCCCGUUUGCCCGCUGUGCGUGUGUGUGUGUG .....((..((...((....))....))..)).........((((((((((((((((((.......(((((((((..((....))..)))))))))))))))))).)))))))))..... ( -45.01) >DroYak_CAF1 336 114 - 1 GUGAAUCAAGCGGAGUGACAACAAAAGCGGGAAAUGUGAAAAUGCGCACGGCAGUGGGUUGAAGCGAGACGGGAAUA------GCGAUGCCCGUUUGCCCGCUGUGCGUGUGUGUGUGUG .....((..((...((....))....))..)).........((((((((((((((((((.......(((((((.((.------...)).)))))))))))))))).)))))))))..... ( -38.11) >consensus AUAAAUCAAGCGAAGUAGCAACAAAAGCGGGAAAUGUGAAAAUGCGCGCGGCAGUGGGUUGAAGCGAGACGGGUAGAGCUAGAGCGAUGCCCGUUUGCCCGCUGUGCGUGUGUGUGUGUG .....((..((...((....))....))..)).........((((((((((((((((((.......(((((((((..((....))..)))))))))))))))))).)))))))))..... (-38.77 = -39.59 + 0.81)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:41:31 2006