
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 25,841,042 – 25,841,167 |
| Length | 125 |
| Max. P | 0.953266 |

| Location | 25,841,042 – 25,841,134 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 92 |
| Reading direction | forward |
| Mean pairwise identity | 84.24 |
| Mean single sequence MFE | -24.06 |
| Consensus MFE | -16.40 |
| Energy contribution | -17.56 |
| Covariance contribution | 1.16 |
| Combinations/Pair | 1.26 |
| Mean z-score | -2.53 |
| Structure conservation index | 0.68 |
| SVM decision value | 1.22 |
| SVM RNA-class probability | 0.932675 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 25841042 92 + 27905053 AGGAAUUUUGAAAUCUAUUGUUUCUUUCCACAUUUUAUUUUGAUAUUUCAAAAACUACCAGCUGCCUGUGUUGAGCAACGCACGGGCAGUUC .((((....(((((.....))))).))))........((((((....))))))......((((((((((((........)))))))))))). ( -27.90) >DroSec_CAF1 12888 92 + 1 UGGAAUUUGGGAAUAUAUUGUUUUUUUCCACAUUUUAUAUUGAUAUUUCAAAAACUACCAGCUGCCUGUGUUGAGCAACGCACGGCCAGUUC ((((((.((((((...........)))))).))))))..((((....))))........(((((((((((((....)))))).)).))))). ( -21.40) >DroSim_CAF1 12981 92 + 1 UGGAAUUUGGGAAUAUAUUGUUUUUUUCCACAUUUCAUAUUGAUAUUUCAAAAACUACCAGCUGCCUGUGUUGAGCAACGCACGGGCAGUUC ((((((.((((((...........)))))).))))))..((((....))))........((((((((((((........)))))))))))). ( -29.10) >DroEre_CAF1 13117 88 + 1 AGGAAUUUUAGGCUAUAUUGU----UUCUACAUUUAAUUGUGAAGUUUCAAAAAUUACCAGCUGCCUGUGUUGAGCAACGCACUGCUAGUUC .(((((((((.(......(((----....)))......).)))))))))..........((((((.((((((....))))))..)).)))). ( -15.80) >DroYak_CAF1 12959 92 + 1 UGAAUUUCUAGGCUAUAUUGUGUAUUUGUACAUUUUAUUUUGAUGUUUCGAAAUUUACCAGCUGCCUGUGUUGAGCAACGCACGGGUGGUUC ((((((((.((((.(((..(((((....)))))..)))......)))).))))))))..(((..(((((((........)))))))..))). ( -26.10) >consensus UGGAAUUUUGGAAUAUAUUGUUUUUUUCCACAUUUUAUUUUGAUAUUUCAAAAACUACCAGCUGCCUGUGUUGAGCAACGCACGGGCAGUUC .(((((.(((((((....(((........)))....))))))).)))))..........((((((((((((........)))))))))))). (-16.40 = -17.56 + 1.16)

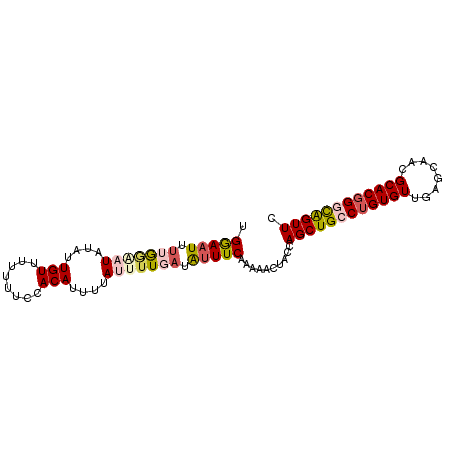
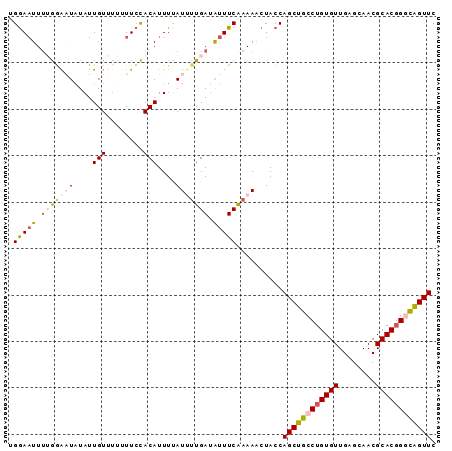
| Location | 25,841,074 – 25,841,167 |
|---|---|
| Length | 93 |
| Sequences | 5 |
| Columns | 94 |
| Reading direction | forward |
| Mean pairwise identity | 84.53 |
| Mean single sequence MFE | -27.30 |
| Consensus MFE | -19.88 |
| Energy contribution | -21.08 |
| Covariance contribution | 1.20 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.49 |
| Structure conservation index | 0.73 |
| SVM decision value | 1.43 |
| SVM RNA-class probability | 0.953266 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 25841074 93 + 27905053 UUUUAUUUUGAUAUUUCAAAAACUACCAGCUGCCUGUGUUGAGCAACGCACGGGCAGUUCGAUAAUACCGGCUAUCGAUAUUUAUCGAUUG-UU .....((((((....))))))....(.((((((((((((........)))))))))))).)........(((.((((((....)))))).)-)) ( -28.60) >DroSec_CAF1 12920 93 + 1 UUUUAUAUUGAUAUUUCAAAAACUACCAGCUGCCUGUGUUGAGCAACGCACGGCCAGUUCGAUAGCACCGGCUACCGGUAGUUAUCGAUUG-UU .......((((....)))).........((((((((((((....)))))).)).))))(((((((((((((...))))).))))))))...-.. ( -30.80) >DroSim_CAF1 13013 93 + 1 UUUCAUAUUGAUAUUUCAAAAACUACCAGCUGCCUGUGUUGAGCAACGCACGGGCAGUUCGAUAGCACCGGCUACCAGUAGUUAUCGAUUG-UU .......((((....)))).........(((((((((((........)))))))))))((((((((((.((...)).)).))))))))...-.. ( -30.30) >DroEre_CAF1 13145 94 + 1 UUUAAUUGUGAAGUUUCAAAAAUUACCAGCUGCCUGUGUUGAGCAACGCACUGCUAGUUCGACAACAUUGGCUAUCGGUAUUUAUCGAUGAACC ..(((((.(((....)))..)))))......(((..((((((((...((...))..)))))))).....)))(((((((....))))))).... ( -18.70) >DroYak_CAF1 12991 94 + 1 UUUUAUUUUGAUGUUUCGAAAUUUACCAGCUGCCUGUGUUGAGCAACGCACGGGUGGUUCGAUAACAACGGCUAUCGGUAUACAUCGAUAGGAC .......(((.((((((((.........((..(((((((........)))))))..)))))).)))).)))((((((((....))))))))... ( -28.10) >consensus UUUUAUUUUGAUAUUUCAAAAACUACCAGCUGCCUGUGUUGAGCAACGCACGGGCAGUUCGAUAACACCGGCUAUCGGUAUUUAUCGAUUG_UU .....((((((....)))))).......(((((((((((........)))))))))))((((((..(((((...)))))...))))))...... (-19.88 = -21.08 + 1.20)



| Location | 25,841,074 – 25,841,167 |
|---|---|
| Length | 93 |
| Sequences | 5 |
| Columns | 94 |
| Reading direction | reverse |
| Mean pairwise identity | 84.53 |
| Mean single sequence MFE | -24.80 |
| Consensus MFE | -19.76 |
| Energy contribution | -18.36 |
| Covariance contribution | -1.40 |
| Combinations/Pair | 1.36 |
| Mean z-score | -2.11 |
| Structure conservation index | 0.80 |
| SVM decision value | 1.18 |
| SVM RNA-class probability | 0.926745 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 25841074 93 - 27905053 AA-CAAUCGAUAAAUAUCGAUAGCCGGUAUUAUCGAACUGCCCGUGCGUUGCUCAACACAGGCAGCUGGUAGUUUUUGAAAUAUCAAAAUAAAA ..-...(((((((.(((((.....))))))))))))((((((.....((((((.......)))))).))))))((((((....))))))..... ( -25.10) >DroSec_CAF1 12920 93 - 1 AA-CAAUCGAUAACUACCGGUAGCCGGUGCUAUCGAACUGGCCGUGCGUUGCUCAACACAGGCAGCUGGUAGUUUUUGAAAUAUCAAUAUAAAA ..-...((((.((((((((((.((((((........))))))(.((.(((....))).)).)..)))))))))).))))............... ( -30.20) >DroSim_CAF1 13013 93 - 1 AA-CAAUCGAUAACUACUGGUAGCCGGUGCUAUCGAACUGCCCGUGCGUUGCUCAACACAGGCAGCUGGUAGUUUUUGAAAUAUCAAUAUGAAA ..-...((((.((((((..((.(((((.((.........)))).((.(((....))).))))).))..)))))).))))............... ( -28.00) >DroEre_CAF1 13145 94 - 1 GGUUCAUCGAUAAAUACCGAUAGCCAAUGUUGUCGAACUAGCAGUGCGUUGCUCAACACAGGCAGCUGGUAAUUUUUGAAACUUCACAAUUAAA ((((..((((.((.(((((...(((..(((((.......(((((....))))))))))..)))...))))).)).))))))))........... ( -19.41) >DroYak_CAF1 12991 94 - 1 GUCCUAUCGAUGUAUACCGAUAGCCGUUGUUAUCGAACCACCCGUGCGUUGCUCAACACAGGCAGCUGGUAAAUUUCGAAACAUCAAAAUAAAA ......((((.((.((((((((((....)))))).............((((((.......)))))).)))).)).))))............... ( -21.30) >consensus AA_CAAUCGAUAAAUACCGAUAGCCGGUGUUAUCGAACUGCCCGUGCGUUGCUCAACACAGGCAGCUGGUAGUUUUUGAAAUAUCAAAAUAAAA ......((((.((.((((((((((....)))))).............((((((.......)))))).)))).)).))))............... (-19.76 = -18.36 + -1.40)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:41:25 2006