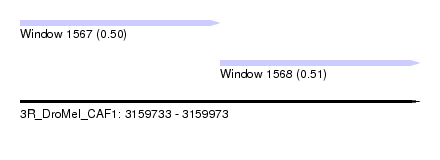
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 3,159,733 – 3,159,973 |
| Length | 240 |
| Max. P | 0.505996 |

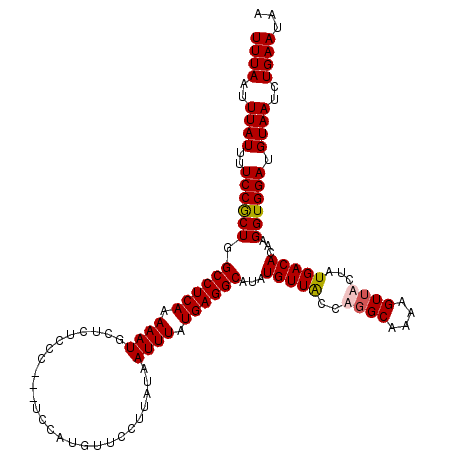
| Location | 3,159,733 – 3,159,853 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.69 |
| Mean single sequence MFE | -24.00 |
| Consensus MFE | -18.77 |
| Energy contribution | -19.03 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.44 |
| Structure conservation index | 0.78 |
| SVM decision value | -0.07 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 3159733 120 + 27905053 GCGUGUCCUUUAAACUUUUAAAGCUUAAGCAUGCGUGGGAAUAAUUAAAAUGUUAAUGCCCCGUACUCAAGGCUUAACCUCAUUUAUAGAAAGUCACCUCAGUAAGUCCUACGUACUUAU ((.((..((((((....))))))..)).)).(((((((((...........(((((.(((..........))))))))....(((((.((........)).))))))))))))))..... ( -24.50) >DroSec_CAF1 7607 120 + 1 GCGUAUACUUUAAACUCUUAAAGCUUAAGCGUGCAUAGGAUUAAUUAAAAUGUUAAUGCCACGUACUCAGGGCUUAACCUCAUUUAUAGAAAGUCACCUCAGUAAAUCCUACGUACUUAU ((.((..((((((....))))))..)).))((((.(((((((.(((.....(((((.(((.(.......)))))))))..........((........))))).))))))).)))).... ( -25.00) >DroSim_CAF1 9791 120 + 1 GCGUAUCCUUUAAACUCUUAAAGCUUAAGCGUGCAUAGGAUUAAUUAAAAUGUUAAUGCCACGUACUCAGGGCUUAACCUCAUUUAUAGAAAGUCACCUCAGUAAAUCCUACGUACUUAU ((.((..((((((....))))))..)).))((((.(((((((.(((.....(((((.(((.(.......)))))))))..........((........))))).))))))).)))).... ( -25.00) >DroYak_CAF1 9153 120 + 1 GCGUAUCCUUUUAACUCUUAAAGCUUAAGCGUGCAUGCGAAUAAUUAAAAUGUUAAUGCUCUGUACUCUACGCUUAACCUCAUUUAUAGGAAGUGCCCCCAGUUAAUCCUACGUACUUAU (((((.....((((((......(((((((((((..((((....(((((....)))))....))))...)))))))..(((.......)))))))......))))))...)))))...... ( -21.50) >consensus GCGUAUCCUUUAAACUCUUAAAGCUUAAGCGUGCAUAGGAAUAAUUAAAAUGUUAAUGCCACGUACUCAAGGCUUAACCUCAUUUAUAGAAAGUCACCUCAGUAAAUCCUACGUACUUAU ((.((..((((((....))))))..)).))((((.(((((...........(((((.(((..........))))))))....(((((.((........)).)))))))))).)))).... (-18.77 = -19.03 + 0.25)

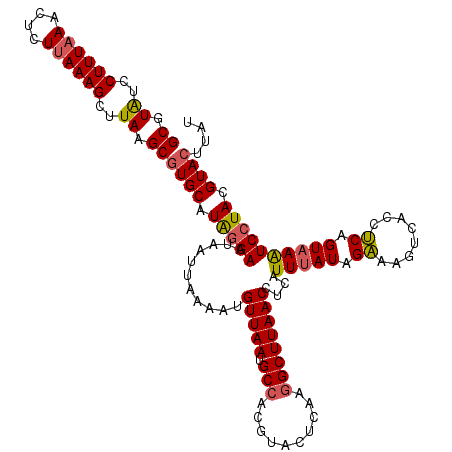
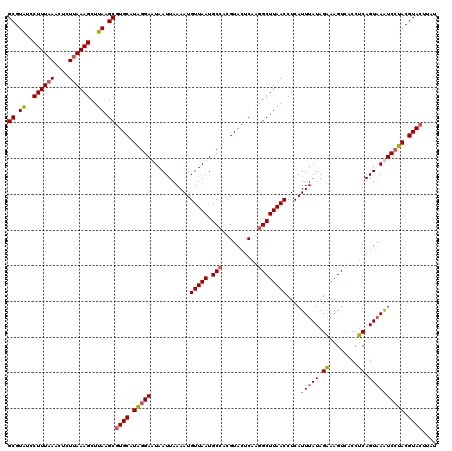
| Location | 3,159,853 – 3,159,973 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.09 |
| Mean single sequence MFE | -23.42 |
| Consensus MFE | -22.17 |
| Energy contribution | -22.55 |
| Covariance contribution | 0.37 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.30 |
| Structure conservation index | 0.95 |
| SVM decision value | -0.05 |
| SVM RNA-class probability | 0.505996 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 3159853 120 + 27905053 UUUAAUUUAUUUUCCGCUGGCCUCAAAAAUGUUCUCCCUCCUCCAUGUUCCUCAUAAUUUAUGAGGCAUAUGUUACCAUGCAAAAGUUACUAUGACAUAAGGUGGAUGUAAUCUGAAUAA ((((..((((..((((((.((((((.((((..........................)))).)))))).(((((((....((....)).....))))))).)))))).))))..))))... ( -24.77) >DroSec_CAF1 7727 117 + 1 UUUAAUUUAUUUUCCGCUGGCCUCAAAAAUGCUCUCCC---UCCAUUUUCCUUAUAAUUUAUGAGGCAUAUGUUGCCUGGCAAAAGUUACUAUGACACAAGGUGGAUGUAAUCUGAAUAA ((((..((((..((((((.(((...((((((.......---..))))))..............(((((.....))))))))....(((.....)))....)))))).))))..))))... ( -22.70) >DroSim_CAF1 9911 117 + 1 UUUAAUUUAUUUUCCGCUGGCCUCAAAAAUGCUCUCCC---UCCAUUUUCCUUAUAAUUUAUGAGGCAUAUGUUACCUGGCAAAAGUUACUAUGACACAAGGUGGAUGUAAUCUGAAUAA ((((..((((..((((((.((((((.((((........---...............)))).))))))...(((((..((((....))))...)))))...)))))).))))..))))... ( -23.60) >DroYak_CAF1 9273 117 + 1 UUUAAUUUAUUUUCCACUGGCCUCAAAAAUGUUCUCCC---UCCCUGUUCCUUAUAAUUUAUGAGGCAUAUGUUACCAGGCAAAAGUUACUAUGACACUAGGUGGAUGUAAUCUGAAUAA ((((..((((..((((((.((((((.((((........---...............)))).))))))...(((((..((..........)).)))))...)))))).))))..))))... ( -22.60) >consensus UUUAAUUUAUUUUCCGCUGGCCUCAAAAAUGCUCUCCC___UCCAUGUUCCUUAUAAUUUAUGAGGCAUAUGUUACCAGGCAAAAGUUACUAUGACACAAGGUGGAUGUAAUCUGAAUAA ((((..((((..((((((.((((((.((((..........................)))).))))))...(((((..((((....))))...)))))...)))))).))))..))))... (-22.17 = -22.55 + 0.37)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:57:10 2006