| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 25,794,971 – 25,795,075 |
| Length | 104 |
| Max. P | 0.813088 |

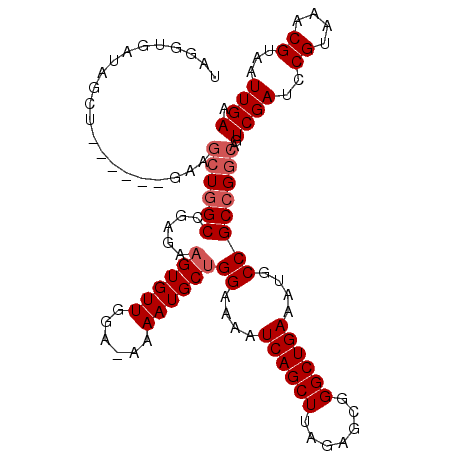
| Location | 25,794,971 – 25,795,075 |
|---|---|
| Length | 104 |
| Sequences | 5 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 87.64 |
| Mean single sequence MFE | -22.76 |
| Consensus MFE | -17.04 |
| Energy contribution | -17.52 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.68 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.21 |
| SVM RNA-class probability | 0.635501 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 25794971 104 + 27905053 UUCAAUUACGUUUACGGAUCGACUGCCGGCGGCAUUUCAGCCAGCUCUAAGCUGAUUUUCCAGCAUUUU-UCCAACACUUCUCCGCCAGCUUC------AGCUAUCACCUA ........((....))(((.(...((.((((((......)))........((((......)))).....-..............))).))...------..).)))..... ( -20.60) >DroSec_CAF1 38338 104 + 1 UUCAAUUACGUUUACGGAUCGACUGCCGGCGGCAUUUCAGCCCGCUCUAAGCUGACUUUCCAGCAUUUU-UCCAACACUUCUCGGCCAGCUUC------AGCUAUCACCUA ...............((.......(((((((((......).)))).....((((......)))).....-............)))).(((...------.)))....)).. ( -22.20) >DroSim_CAF1 38700 104 + 1 UUCAAUUACGUUUACGGAUCGACUGCCGGCGGCAUUUCAGCCCGCUCUAAGCUGACUUUCCAGCAUUUU-UCCAACACUUCUCGGCCAGCUUC------AGCUAUCACCUA ...............((.......(((((((((......).)))).....((((......)))).....-............)))).(((...------.)))....)).. ( -22.20) >DroEre_CAF1 39857 110 + 1 UUCAAUUACGA-UACGGAUCGACUGCCGGCGGCAUUUCAGCCAGUUCUAAGCUGAUUUUCCAGCAUUUUUUCCAACACUUCUUCGCCAGCUUCAGCUUCUGCUAUCACCUA .........((-((((((......((.((((((......)))........((((......))))....................))).)).......)))).))))..... ( -26.02) >DroYak_CAF1 38160 101 + 1 UUCAAUUACGA-UACGGUUCGACAGCCGGCGGCAUUUCAGCCGAGUCUAAGCUGAUUUUCCGGCAUUUUUUCAAACACUUCUUUGCUA---------UCACCUAUCACCUA .........((-((.(((......(((((((((......))))((((......))))..))))).......((((......))))...---------..)))))))..... ( -22.80) >consensus UUCAAUUACGUUUACGGAUCGACUGCCGGCGGCAUUUCAGCCAGCUCUAAGCUGAUUUUCCAGCAUUUU_UCCAACACUUCUCCGCCAGCUUC______AGCUAUCACCUA ...............((..........((((((......)))........((((......))))....................)))((((........))))....)).. (-17.04 = -17.52 + 0.48)

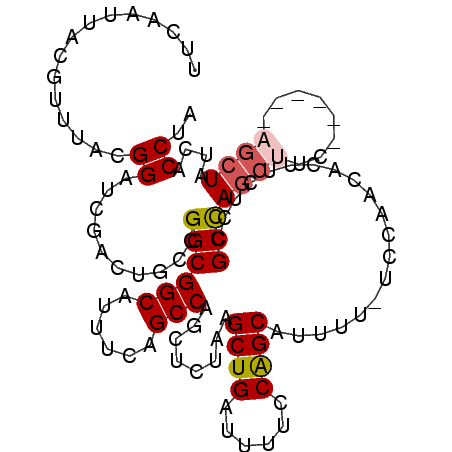
| Location | 25,794,971 – 25,795,075 |
|---|---|
| Length | 104 |
| Sequences | 5 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 87.64 |
| Mean single sequence MFE | -30.72 |
| Consensus MFE | -23.00 |
| Energy contribution | -23.80 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.97 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.65 |
| SVM RNA-class probability | 0.813088 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 25794971 104 - 27905053 UAGGUGAUAGCU------GAAGCUGGCGGAGAAGUGUUGGA-AAAAUGCUGGAAAAUCAGCUUAGAGCUGGCUGAAAUGCCGCCGGCAGUCGAUCCGUAAACGUAAUUGAA ..(((....)))------...((((((((...((((((...-..))))))......((((((.......))))))....))))))))..(((((.((....))..))))). ( -30.80) >DroSec_CAF1 38338 104 - 1 UAGGUGAUAGCU------GAAGCUGGCCGAGAAGUGUUGGA-AAAAUGCUGGAAAGUCAGCUUAGAGCGGGCUGAAAUGCCGCCGGCAGUCGAUCCGUAAACGUAAUUGAA ..((((((.(((------(((((((((.....((((((...-..)))))).....))))))))...((((.(......))))))))).))))..))............... ( -31.50) >DroSim_CAF1 38700 104 - 1 UAGGUGAUAGCU------GAAGCUGGCCGAGAAGUGUUGGA-AAAAUGCUGGAAAGUCAGCUUAGAGCGGGCUGAAAUGCCGCCGGCAGUCGAUCCGUAAACGUAAUUGAA ..((((((.(((------(((((((((.....((((((...-..)))))).....))))))))...((((.(......))))))))).))))..))............... ( -31.50) >DroEre_CAF1 39857 110 - 1 UAGGUGAUAGCAGAAGCUGAAGCUGGCGAAGAAGUGUUGGAAAAAAUGCUGGAAAAUCAGCUUAGAACUGGCUGAAAUGCCGCCGGCAGUCGAUCCGUA-UCGUAAUUGAA ..(((((((((....)))...((((((.....((((((......))))))((....((((((.......))))))....)))))))).))))..))...-........... ( -30.90) >DroYak_CAF1 38160 101 - 1 UAGGUGAUAGGUGA---------UAGCAAAGAAGUGUUUGAAAAAAUGCCGGAAAAUCAGCUUAGACUCGGCUGAAAUGCCGCCGGCUGUCGAACCGUA-UCGUAAUUGAA .....(((((((((---------((((......((((((....))))))(((....((((((.......))))))....)))...))))))..))).))-))......... ( -28.90) >consensus UAGGUGAUAGCU______GAAGCUGGCCGAGAAGUGUUGGA_AAAAUGCUGGAAAAUCAGCUUAGAGCGGGCUGAAAUGCCGCCGGCAGUCGAUCCGUAAACGUAAUUGAA .....................((((((.....((((((......))))))((....((((((.......))))))....))))))))..((((..((....))...)))). (-23.00 = -23.80 + 0.80)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:41:15 2006