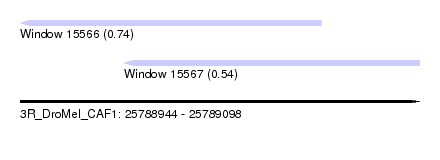
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 25,788,944 – 25,789,098 |
| Length | 154 |
| Max. P | 0.740442 |

| Location | 25,788,944 – 25,789,060 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.90 |
| Mean single sequence MFE | -32.13 |
| Consensus MFE | -22.95 |
| Energy contribution | -24.41 |
| Covariance contribution | 1.46 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.45 |
| SVM RNA-class probability | 0.740442 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
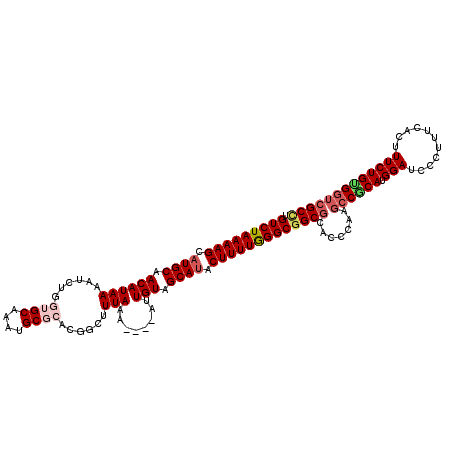
>3R_DroMel_CAF1 25788944 116 - 27905053 UGUCUAAAAGCAUGCAACAUAAAAUGUGGUGCAAAUGCGCACGGCUUUAAA----AUUGUAGCAUACUUUUGGGCUGCGCACCCAAGCCGCAUGGAUCCCUUUCACUUUCUGCGGUCGCC .(((((((((.((((.(((......((.((((......)))).))......----..))).)))).))))))))).(((.......((((((.(((...........)))))))))))). ( -36.75) >DroSec_CAF1 32416 115 - 1 UGUCUAAAAGCAUGCAACAUAAAAUUUGGUGCAAAUGCGCACGGCUUUAAA----AGUGUAGCAUACUUUUGGGCGGCGCACCCAAGCCGCAUGGAUC-CUUUCACCUUCUGUGGUCGCC .(((((((((.((((.((((.(((.(((((((....)))).))).)))...----.)))).)))).)))))))))((((.......((((((((((..-..)))).....)))))))))) ( -35.51) >DroSim_CAF1 32688 114 - 1 UGUCUAAAAGCAUGCAACAUAAAAUUUGGUGCAAAUGCGCACGGCUUUAA-----AUUGUAGCAUACUUUUGGGCGGCACAGCCAAGCCGCAUGGAUC-CUUUCACCUUCUGUGGUCGCC .(((((((((.((((.(((..(((.(((((((....)))).))).)))..-----..))).)))).)))))))))(((...)))..((((((((((..-..)))).....)))))).... ( -33.70) >DroEre_CAF1 33879 119 - 1 AGUCUAAAAGCAUGCAACAUAAUAUCCACAGCGAAUGCGCACGACUUUA-AAUAUACUGUAGCAUACUUUUAGGCGGCGCACCCAAGCCACACGGAUUCCUUUCACUUUCUGUGGUCGCU .(((((((((.((((.(((..((((....((((........)).))...-.))))..))).)))).)))))))))(((........))).((((((............))))))...... ( -30.80) >DroYak_CAF1 31867 108 - 1 UGUCUAAAAGCAUGCAACAUAAUAUC------------GCACGACUUUAAAAUAUAUUGUAGCACACUUUUAGGCGGCGCACCCAUUACACAAGGAUUCCUUUCACUUUCUGCGGUCGCU .(((((((((..(((.(((..((((.------------.............))))..))).)))..)))))))))((((.(((((......((((...))))........)).))))))) ( -23.88) >consensus UGUCUAAAAGCAUGCAACAUAAAAUCUGGUGCAAAUGCGCACGGCUUUAAA____AUUGUAGCAUACUUUUGGGCGGCGCACCCAAGCCGCAUGGAUCCCUUUCACUUUCUGUGGUCGCC .(((((((((.((((.((((((......((((....))))......)))........))).)))).)))))))))((((.......((((((.(((...........))))))))))))) (-22.95 = -24.41 + 1.46)

| Location | 25,788,984 – 25,789,098 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 79.78 |
| Mean single sequence MFE | -28.48 |
| Consensus MFE | -16.58 |
| Energy contribution | -17.84 |
| Covariance contribution | 1.26 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.58 |
| SVM decision value | 0.02 |
| SVM RNA-class probability | 0.543720 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
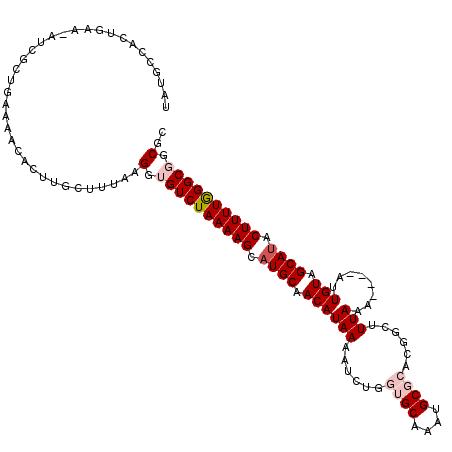
>3R_DroMel_CAF1 25788984 114 - 27905053 GAUGAAACUAAAUAUCACUGAAAACACUUGCAUUAAGGUGUCUAAAAGCAUGCAACAUAAAAUGUGGUGCAAAUGCGCACGGCUUUAAA----AUUGUAGCAUACUUUUGGGCUGCGC ((((........)))).........((((......))))(((((((((.((((.(((......((.((((......)))).))......----..))).)))).)))))))))..... ( -27.74) >DroSec_CAF1 32455 113 - 1 UAUCGCACUGAA-AUCGCUGAAAACACUUGCUUUAAGGUGUCUAAAAGCAUGCAACAUAAAAUUUGGUGCAAAUGCGCACGGCUUUAAA----AGUGUAGCAUACUUUUGGGCGGCGC ....((((..((-...(((....((((((......)))))).....)))(((...)))....))..))))....((((.((.((..(((----(((((...)))))))))).)))))) ( -32.30) >DroSim_CAF1 32727 112 - 1 UAUCGCACUAAA-AUCGCUGCAAACACUUUCUUUAAGGUGUCUAAAAGCAUGCAACAUAAAAUUUGGUGCAAAUGCGCACGGCUUUAA-----AUUGUAGCAUACUUUUGGGCGGCAC ............-...(((((..(((((((....)))))))(((((((.((((.(((..(((.(((((((....)))).))).)))..-----..))).)))).)))))))))))).. ( -33.10) >DroEre_CAF1 33919 116 - 1 UAUGCCACAGAA-AUCGAUGAAAACUCUGGCUUUGAGGAGUCUAAAAGCAUGCAACAUAAUAUCCACAGCGAAUGCGCACGACUUUA-AAUAUACUGUAGCAUACUUUUAGGCGGCGC ...(((......-...........(((.......)))..(((((((((.((((.(((..((((....((((........)).))...-.))))..))).)))).)))))))))))).. ( -27.30) >DroYak_CAF1 31907 105 - 1 UAUGCCACAGAA-AUCGAUGAACACACUGGCACGUAGGUGUCUAAAAGCAUGCAACAUAAUAUC------------GCACGACUUUAAAAUAUAUUGUAGCACACUUUUAGGCGGCGC ..(((((.....-..............)))))....(.((((((((((..(((.(((..((((.------------.............))))..))).)))..)))))))))).).. ( -21.95) >consensus UAUGCCACUGAA_AUCGCUGAAAACACUUGCUUUAAGGUGUCUAAAAGCAUGCAACAUAAAAUCUGGUGCAAAUGCGCACGGCUUUAAA____AUUGUAGCAUACUUUUGGGCGGCGC ....................................(.((((((((((.((((.((((((......((((....))))......)))........))).)))).)))))))))).).. (-16.58 = -17.84 + 1.26)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:41:13 2006