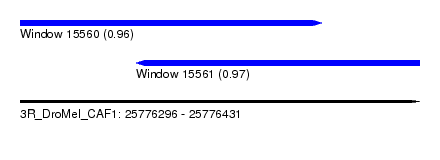
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 25,776,296 – 25,776,431 |
| Length | 135 |
| Max. P | 0.970013 |

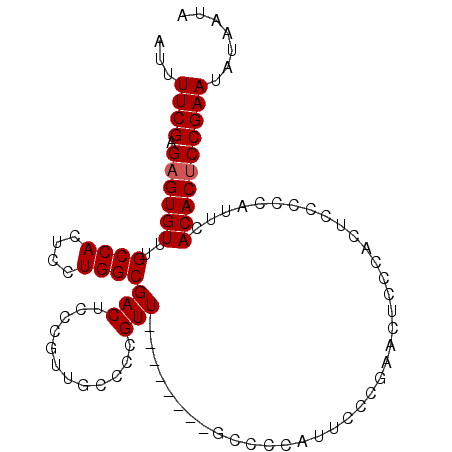
| Location | 25,776,296 – 25,776,398 |
|---|---|
| Length | 102 |
| Sequences | 4 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 86.73 |
| Mean single sequence MFE | -18.96 |
| Consensus MFE | -13.68 |
| Energy contribution | -13.92 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.86 |
| Structure conservation index | 0.72 |
| SVM decision value | 1.51 |
| SVM RNA-class probability | 0.960395 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 25776296 102 + 27905053 AUUUUCGAGAGUGUUUGCCACUCCUGGCGACUCCCGUUGCCCCGUUGCCCGGUUGCCCCAUUCCCCAACUCCCAUUCCCCCAUUCACACUCCGAAUAUAAUA ...((((.((((((..((.((....((((((....))))))..)).))..(((((..........)))))...............))))))))))....... ( -22.90) >DroSec_CAF1 19899 94 + 1 AUUUUCGAGAGUGUUUGCCACUCCUGGCGACUCUCUUUGCCCCGUU--------GCCCCAUUCCCGAACUCCCACUCCCCCAUUUACACUCCGAAUAUAAUA ...((((.((((((((((((....))))))........((......--------)).............................))))))))))....... ( -17.60) >DroSim_CAF1 20064 94 + 1 AUUUUCGAGAGUGUUUGCCACUCCUGGCGACUCUCGUUGCCCCGUU--------GCCCCAUUCCCGAACUCCCACUCCCCCAUUCACACUCCGAAUAUAAUA ...((((.((((((..((.((....((((((....))))))..)).--------)).........(((..............)))))))))))))....... ( -20.04) >DroYak_CAF1 18755 87 + 1 AUUUUCGAGUGUGUUUGCCACUCCUGGCGACUCCCAUU-CCCCGUU--------CCCCCAUUUCU------CCGUUCCCCCAUUCACACUCCGAAUAUAAUA ...((((((((((.((((((....))))))........-....(..--------....)......------.............)))))).))))....... ( -15.30) >consensus AUUUUCGAGAGUGUUUGCCACUCCUGGCGACUCCCGUUGCCCCGUU________GCCCCAUUCCCGAACUCCCACUCCCCCAUUCACACUCCGAAUAUAAUA ...((((.((((((..((((....))))(((............))).......................................))))))))))....... (-13.68 = -13.92 + 0.25)


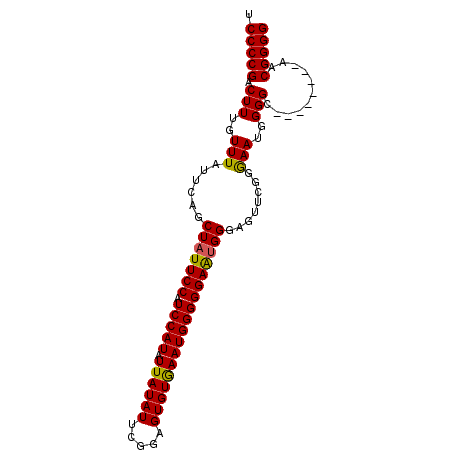
| Location | 25,776,335 – 25,776,431 |
|---|---|
| Length | 96 |
| Sequences | 4 |
| Columns | 96 |
| Reading direction | reverse |
| Mean pairwise identity | 88.59 |
| Mean single sequence MFE | -30.75 |
| Consensus MFE | -25.84 |
| Energy contribution | -25.46 |
| Covariance contribution | -0.38 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.40 |
| Structure conservation index | 0.84 |
| SVM decision value | 1.65 |
| SVM RNA-class probability | 0.970013 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 25776335 96 - 27905053 UCCCCGACUUUGUUUAUUCAGCUAUUCCAUCCAUAUUAUAUUCGGAGUGUGAAUGGGGGAAUGGGAGUUGGGGAAUGGGGCAACCGGGCAACGGGG ((((((((((((......)).(((((((.(((((.((((((.....)))))))))))))))))))))))))))).........((.(....).)). ( -37.60) >DroSec_CAF1 19938 88 - 1 UCCCCGACUUUGUUUAUUCAGCUAUUCCAUCCAUAUUAUAUUCGGAGUGUAAAUGGGGGAGUGGGAGUUCGGGAAUGGGGC--------AACGGGG .(((((.(((..(((((((..(((((((.(((((.((((((.....))))))))))))))))))))))....)))..))).--------..))))) ( -29.20) >DroSim_CAF1 20103 88 - 1 UCCCCGACUUUGUUUAUUCAGCUAUUCCAUCCAUAUUAUAUUCGGAGUGUGAAUGGGGGAGUGGGAGUUCGGGAAUGGGGC--------AACGGGG .(((((.(((..(((((((..(((((((.(((((.((((((.....))))))))))))))))))))))....)))..))).--------..))))) ( -29.30) >DroYak_CAF1 18793 82 - 1 UCCCCGACUUUGUUUAUUCAGCUAUUCCAUCCAUAUUAUAUUCGGAGUGUGAAUGGGGGAACGG------AGAAAUGGGGG--------AACGGGG .(((((.(((..(((......(..((((.(((((.((((((.....)))))))))))))))..)------..)))..))).--------..))))) ( -26.90) >consensus UCCCCGACUUUGUUUAUUCAGCUAUUCCAUCCAUAUUAUAUUCGGAGUGUGAAUGGGGGAAUGGGAGUUCGGGAAUGGGGC________AACGGGG .(((((.(((..(((......(((((((.(((((.((((((.....))))))))))))))))))........)))..)))...........))))) (-25.84 = -25.46 + -0.38)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:41:07 2006