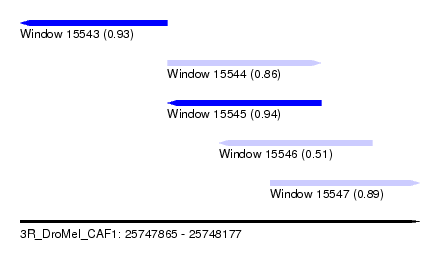
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 25,747,865 – 25,748,177 |
| Length | 312 |
| Max. P | 0.942927 |

| Location | 25,747,865 – 25,747,980 |
|---|---|
| Length | 115 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 77.26 |
| Mean single sequence MFE | -26.75 |
| Consensus MFE | -16.66 |
| Energy contribution | -18.58 |
| Covariance contribution | 1.92 |
| Combinations/Pair | 1.30 |
| Mean z-score | -1.98 |
| Structure conservation index | 0.62 |
| SVM decision value | 1.21 |
| SVM RNA-class probability | 0.931651 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 25747865 115 - 27905053 AAUACGAACAGUUUCAUCUUGGUCAAGUCUUGCAAC---UCAAUGGUGUUCCGAUGUCGAAGCGCCUUUAUAUACAGAAGAA--GUCCCUUGUCUUAUCUUGACCAAGUGUCAAACUACC .........(((((.((((((((((((....((...---.....((((((.((....)).))))))...........(((..--....))))).....)))))))))).)).)))))... ( -27.80) >DroSec_CAF1 19150 115 - 1 AAUACGAACAGUUUCAUCUUGGUCAAGUCUUGCAAC---UCAAUGGUGUUCCGAGUCCGAAGCGCCUUUAUAUACAGAAGAA--GUCCCUUGUCUUAUCUUGACCAAGUGUCAAACUACC .........(((((.((((((((((((....((...---.....((((((.((....)).))))))...........(((..--....))))).....)))))))))).)).)))))... ( -27.20) >DroSim_CAF1 12924 115 - 1 AAUACGAACAGUUUCAUCUUGGUCAAGUCUUGCAAC---UCAAUGGUGUUCCGAAUCCGAAGCGCCUUUAUAUACAGAAGAA--GUCCCUUGUCUUAUCUUGACCAAGUGUCAAACUACC .........(((((.((((((((((((....((...---.....((((((.((....)).))))))...........(((..--....))))).....)))))))))).)).)))))... ( -27.20) >DroEre_CAF1 12921 115 - 1 AAUACGAAUAGUUUCAUCUUGGUCAAGUCUUGCAAC---UUAAUGGUGUUCUGAAACCAAGGCGCCCUUUUAUACAAAAGAA--AUCCGCUGUCUUAUCUUGGCCAAGAGCCAAACUACA ........((((((..(((((((((((....(((.(---.....(((((..((....))..)))))(((((....)))))..--....).))).....)))))))))))...)))))).. ( -32.10) >DroWil_CAF1 28136 100 - 1 AAUAAGAACGAUUUCAUCUUGAGCAACUUGAACAACUACUCAUGGUUUUUCCUAAACUCUCCCACCUUUUUAUACAAAACAGUGAACGC-----UUAUCUC---UGAG------------ .............(((...(((((.....(((.(((((....))))).)))...........(((..((((....))))..)))...))-----)))....---))).------------ ( -11.40) >DroYak_CAF1 13225 115 - 1 AAUACGAACAGUUUCAUCUUGGUCAAGUCUUGCAAC---UCAAUGGUGUUCCGAAACCGAAGCGCCUUUUUAUACCAAAGGA--AUCCGUUGUCUUAUCUUGACCAAGAGCUGAACUACG .........(((((..(((((((((((....(((((---.....((((((.((....)).)))))).............((.--..))))))).....)))))))))))...)))))... ( -34.80) >consensus AAUACGAACAGUUUCAUCUUGGUCAAGUCUUGCAAC___UCAAUGGUGUUCCGAAACCGAAGCGCCUUUAUAUACAAAAGAA__AUCCCUUGUCUUAUCUUGACCAAGUGUCAAACUACC .........(((((...((((((((((.................((((((.((....)).))))))...........((((...........))))..))))))))))....)))))... (-16.66 = -18.58 + 1.92)



| Location | 25,747,980 – 25,748,100 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.94 |
| Mean single sequence MFE | -48.87 |
| Consensus MFE | -42.17 |
| Energy contribution | -41.95 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.81 |
| SVM RNA-class probability | 0.855705 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 25747980 120 + 27905053 CCUGGCCUUGGCCGUGGCCGCAGCUACUGCUGUGCCCGCUCCCGCCCAGAAGCUGACCCCGACGCCCAUCAAGGACAUUCAGGGUCGCAUCACCAACGGCUACCCAGCCUACGAGGGCAA ...(((....)))((((.((((((....)))))).))))....((((.((.((.(((((.((..((......))....)).))))))).))......((((....)))).....)))).. ( -50.90) >DroSec_CAF1 19265 120 + 1 CCUGGCCUUGGCCGUGGCCGCAGCCACUGCUGUGCCCGCUCCCGCCCAGAAGCUGACCCCGACGCCCAUCAAGGACAUCCAGGGUCGCAUCACCAACGGCUACCCAGCCUGCGAGGGCAA ...(((....)))((((.((((((....)))))).))))....((((.((.((.(((((.((.(.((.....)).).))..))))))).))......((((....)))).....)))).. ( -49.30) >DroSim_CAF1 13039 120 + 1 CCUGGCCUUGGCCGUGGCCGCAGCCACUGCUGUGCCCGCUCCCGCCCAGAAGCUGACCCCGACGCCCAUCAAGGACAUCCAGGGUCGCAUCACCAACGGCUACCCAGCCUACGAGGGCAA ...(((....)))((((.((((((....)))))).))))....((((.((.((.(((((.((.(.((.....)).).))..))))))).))......((((....)))).....)))).. ( -49.30) >DroEre_CAF1 13036 120 + 1 CCUGGCCUUGGCCGUGGCCGCAGCCACCGCUGUGCCCGCUCCCACCCAGAAGCUGACCCCGACGCCCAUCAAGGACAUCCAGGGUCGCAUCACCAACGGCUACCCAGCCUACGAGGGCAA ..(((((((((..((((.((((((....)))))).)))).........((.((.(((((.((.(.((.....)).).))..))))))).)).)))).)))))....((((....)))).. ( -45.80) >DroWil_CAF1 28236 117 + 1 CCUGGUCUUGGCUGUGGCCGCAGUCACCGCUGUCCCCACCC---UGCAGAAGCAGAAGCCGGUGCCCGUCAAGGACAUCGAGGGCCGUAUUACCAACGGCUACCCAGCCUAUGAGGGCAA .......((((((((((..(((((....)))))..)))).(---(((....)))).))))))(((((.((((((........((((((.......))))))......))).)))))))). ( -45.44) >DroYak_CAF1 13340 120 + 1 CCUGGCCUUGGCCGUGGCCGCAGCCACUGCUGUGCCCGCUCCCGCCCAGAAGCUGACCCCGACGCCCGUCAAGGACAUCCAGGGUCGCAUCACCAACGGCUACCCGGCCUACGAGGGCAA ...(((....)))((((.((((((....)))))).))))....((((.((.((.(((((.(((....)))..((....)).))))))).))......((((....)))).....)))).. ( -52.50) >consensus CCUGGCCUUGGCCGUGGCCGCAGCCACUGCUGUGCCCGCUCCCGCCCAGAAGCUGACCCCGACGCCCAUCAAGGACAUCCAGGGUCGCAUCACCAACGGCUACCCAGCCUACGAGGGCAA ...(((....)))((((.((((((....)))))).))))....((((.((.((.(((((.((.(.((.....)).).))..))))))).))......((((....)))).....)))).. (-42.17 = -41.95 + -0.22)



| Location | 25,747,980 – 25,748,100 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.94 |
| Mean single sequence MFE | -60.77 |
| Consensus MFE | -53.61 |
| Energy contribution | -54.53 |
| Covariance contribution | 0.92 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.88 |
| SVM decision value | 1.33 |
| SVM RNA-class probability | 0.942927 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 25747980 120 - 27905053 UUGCCCUCGUAGGCUGGGUAGCCGUUGGUGAUGCGACCCUGAAUGUCCUUGAUGGGCGUCGGGGUCAGCUUCUGGGCGGGAGCGGGCACAGCAGUAGCUGCGGCCACGGCCAAGGCCAGG ....(((.((.((((..(((((..(((.((.(((((((((((.(((((.....))))))))))))).((((((....))))))..))))).)))..)))))))))))(((....)))))) ( -61.00) >DroSec_CAF1 19265 120 - 1 UUGCCCUCGCAGGCUGGGUAGCCGUUGGUGAUGCGACCCUGGAUGUCCUUGAUGGGCGUCGGGGUCAGCUUCUGGGCGGGAGCGGGCACAGCAGUGGCUGCGGCCACGGCCAAGGCCAGG .(((....)))((((..((((((((((.((.((((((((((.((((((.....))))))))))))).((((((....))))))..))))).))))))))))(((....)))..))))... ( -64.50) >DroSim_CAF1 13039 120 - 1 UUGCCCUCGUAGGCUGGGUAGCCGUUGGUGAUGCGACCCUGGAUGUCCUUGAUGGGCGUCGGGGUCAGCUUCUGGGCGGGAGCGGGCACAGCAGUGGCUGCGGCCACGGCCAAGGCCAGG ....(((.((.((((..((((((((((.((.((((((((((.((((((.....))))))))))))).((((((....))))))..))))).))))))))))))))))(((....)))))) ( -63.90) >DroEre_CAF1 13036 120 - 1 UUGCCCUCGUAGGCUGGGUAGCCGUUGGUGAUGCGACCCUGGAUGUCCUUGAUGGGCGUCGGGGUCAGCUUCUGGGUGGGAGCGGGCACAGCGGUGGCUGCGGCCACGGCCAAGGCCAGG ....(((.((.((((..(((((((..(.((.((((((((((.((((((.....))))))))))))).((((((....))))))..))))).)..)))))))))))))(((....)))))) ( -63.60) >DroWil_CAF1 28236 117 - 1 UUGCCCUCAUAGGCUGGGUAGCCGUUGGUAAUACGGCCCUCGAUGUCCUUGACGGGCACCGGCUUCUGCUUCUGCA---GGGUGGGGACAGCGGUGACUGCGGCCACAGCCAAGACCAGG ...........((((((((.(((((((........((((((((.....)))).)))).((.(((.((((....)))---)))).))..))))))).......))).)))))......... ( -46.40) >DroYak_CAF1 13340 120 - 1 UUGCCCUCGUAGGCCGGGUAGCCGUUGGUGAUGCGACCCUGGAUGUCCUUGACGGGCGUCGGGGUCAGCUUCUGGGCGGGAGCGGGCACAGCAGUGGCUGCGGCCACGGCCAAGGCCAGG ...........((((..((((((((((.((.((((((((((.((((((.....))))))))))))).((((((....))))))..))))).))))))))))(((....)))..))))... ( -65.20) >consensus UUGCCCUCGUAGGCUGGGUAGCCGUUGGUGAUGCGACCCUGGAUGUCCUUGAUGGGCGUCGGGGUCAGCUUCUGGGCGGGAGCGGGCACAGCAGUGGCUGCGGCCACGGCCAAGGCCAGG ...........((((..((((((((((.((.((((((((((.((((((.....))))))))))))).((((((....))))))..))))).))))))))))(((....)))..))))... (-53.61 = -54.53 + 0.92)

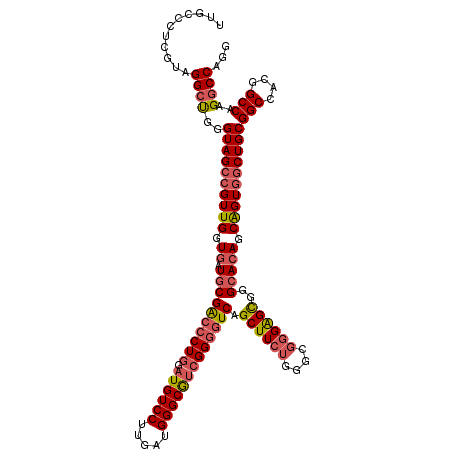
| Location | 25,748,020 – 25,748,140 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.22 |
| Mean single sequence MFE | -54.32 |
| Consensus MFE | -45.78 |
| Energy contribution | -47.75 |
| Covariance contribution | 1.97 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.56 |
| Structure conservation index | 0.84 |
| SVM decision value | -0.06 |
| SVM RNA-class probability | 0.505212 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 25748020 120 - 27905053 UCCGUUGCCGCUGAAGAGCAGACCCACGAUGUAAGGCACCUUGCCCUCGUAGGCUGGGUAGCCGUUGGUGAUGCGACCCUGAAUGUCCUUGAUGGGCGUCGGGGUCAGCUUCUGGGCGGG .......(((((.(..(((.....(((.(.....(((((((.(((......))).)))).)))..).)))....((((((((.(((((.....))))))))))))).)))..).))))). ( -55.80) >DroSec_CAF1 19305 120 - 1 UCCGUUGCCGCUGAAGAGCAGACCCACGAUGUAGGGAACCUUGCCCUCGCAGGCUGGGUAGCCGUUGGUGAUGCGACCCUGGAUGUCCUUGAUGGGCGUCGGGGUCAGCUUCUGGGCGGG .......(((((.(..(((.(((((......(((((....((((..((((.((((....))))....)))).)))))))))(((((((.....))))))).))))).)))..).))))). ( -54.40) >DroSim_CAF1 13079 120 - 1 UCCGUUGCCGCUGAAGAGCAGACCCACGAUGUAGGGAACCUUGCCCUCGUAGGCUGGGUAGCCGUUGGUGAUGCGACCCUGGAUGUCCUUGAUGGGCGUCGGGGUCAGCUUCUGGGCGGG .......(((((.(..(((.(((((......(((((.((((.(((......))).))))...(((.......))).)))))(((((((.....))))))).))))).)))..).))))). ( -54.30) >DroEre_CAF1 13076 120 - 1 UCCGUUGCCGCUGAAGAGCAGACCCACGAUGUAGGGCACCUUGCCCUCGUAGGCUGGGUAGCCGUUGGUGAUGCGACCCUGGAUGUCCUUGAUGGGCGUCGGGGUCAGCUUCUGGGUGGG .......(((((.(..(((..(((.((.(((.(((((.....)))))))).((((....)))))).))).....(((((((.((((((.....))))))))))))).)))..).))))). ( -56.20) >DroWil_CAF1 28276 117 - 1 ACCGUUGCCGCUGAAGCCCAGACCAACGAUGUAGGGCACUUUGCCCUCAUAGGCUGGGUAGCCGUUGGUAAUACGGCCCUCGAUGUCCUUGACGGGCACCGGCUUCUGCUUCUGCA---G .....(((.((.((((((...((((((.(((.(((((.....)))))))).((((....))))))))))......((((((((.....)))).))))...)))))).))....)))---. ( -48.80) >DroYak_CAF1 13380 120 - 1 UCCGUUGCCGCUGAAGAGCAGACCCACGAUGUAGGGCACCUUGCCCUCGUAGGCCGGGUAGCCGUUGGUGAUGCGACCCUGGAUGUCCUUGACGGGCGUCGGGGUCAGCUUCUGGGCGGG .......(((((.(..(((..(((.((.(((.(((((.....)))))))).(((......))))).))).....(((((((.((((((.....))))))))))))).)))..).))))). ( -56.40) >consensus UCCGUUGCCGCUGAAGAGCAGACCCACGAUGUAGGGCACCUUGCCCUCGUAGGCUGGGUAGCCGUUGGUGAUGCGACCCUGGAUGUCCUUGAUGGGCGUCGGGGUCAGCUUCUGGGCGGG .......(((((((((.(((.(((.((.(((.(((((.....)))))))).(((......))))).)))..)))(((((((.((((((.....)))))))))))))..))))..))))). (-45.78 = -47.75 + 1.97)


| Location | 25,748,060 – 25,748,177 |
|---|---|
| Length | 117 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.62 |
| Mean single sequence MFE | -50.28 |
| Consensus MFE | -43.02 |
| Energy contribution | -42.88 |
| Covariance contribution | -0.14 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.99 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.94 |
| SVM RNA-class probability | 0.885702 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 25748060 117 + 27905053 AGGGUCGCAUCACCAACGGCUACCCAGCCUACGAGGGCAAGGUGCCUUACAUCGUGGGUCUGCUCUUCAGCGGCAACGGAAAC---UGGUGGUGCGGUGGCUCGAUCAUCGGCAACACCU .((((((((((((((..(((.(((..((((....))))..)))))).....((((..(.(((((....)))))).))))....---))))))))))((.((.((.....)))).)))))) ( -48.10) >DroSec_CAF1 19345 117 + 1 AGGGUCGCAUCACCAACGGCUACCCAGCCUGCGAGGGCAAGGUUCCCUACAUCGUGGGUCUGCUCUUCAGCGGCAACGGAAAC---UGGUGGUGCGGUGGCUCGAUCAUCGGCAACACCU .((((((((((((((..((..(((..((((....))))..)))..))....((((..(.(((((....)))))).))))....---))))))))))((.((.((.....)))).)))))) ( -45.70) >DroSim_CAF1 13119 117 + 1 AGGGUCGCAUCACCAACGGCUACCCAGCCUACGAGGGCAAGGUUCCCUACAUCGUGGGUCUGCUCUUCAGCGGCAACGGAAAC---UGGUGGUGCGGUGGCUCGAUCAUCGGCAACACCU .((((((((((((((..((..(((..((((....))))..)))..))....((((..(.(((((....)))))).))))....---))))))))))((.((.((.....)))).)))))) ( -45.40) >DroEre_CAF1 13116 117 + 1 AGGGUCGCAUCACCAACGGCUACCCAGCCUACGAGGGCAAGGUGCCCUACAUCGUGGGUCUGCUCUUCAGCGGCAACGGAAAC---UGGUGGUGCGGUGGCUCGAUCAUCGGCAACACCU .((((((((((((((...(((...((((((((((((((.....))))....))))))).)))......)))(....)(....)---))))))))))((.((.((.....)))).)))))) ( -49.10) >DroWil_CAF1 28313 120 + 1 AGGGCCGUAUUACCAACGGCUACCCAGCCUAUGAGGGCAAAGUGCCCUACAUCGUUGGUCUGGGCUUCAGCGGCAACGGUGGCGGUUGGUGGUGCGGUGGUUCCAUCAUUGGCAACACCU (((((((((((((((((.((..(((((((.((((((((.....))))).)))....)).)))))...((.((....)).)))).)))))))))))))).((((((....))).))).))) ( -61.30) >DroYak_CAF1 13420 117 + 1 AGGGUCGCAUCACCAACGGCUACCCGGCCUACGAGGGCAAGGUGCCCUACAUCGUGGGUCUGCUCUUCAGCGGCAACGGAAAC---UGGUGGUGCGGUGGCUCGAUUAUCGGCAACACCU .((((((((((((((...(((....(((((((((((((.....))))....))))))))).(((....))))))...(....)---))))))))))((.((.((.....)))).)))))) ( -52.10) >consensus AGGGUCGCAUCACCAACGGCUACCCAGCCUACGAGGGCAAGGUGCCCUACAUCGUGGGUCUGCUCUUCAGCGGCAACGGAAAC___UGGUGGUGCGGUGGCUCGAUCAUCGGCAACACCU .((((((((((((((.........((((((((((((((.....))))....))))))).)))........((....))........))))))))))((.((.((.....)))).)))))) (-43.02 = -42.88 + -0.14)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:40:54 2006