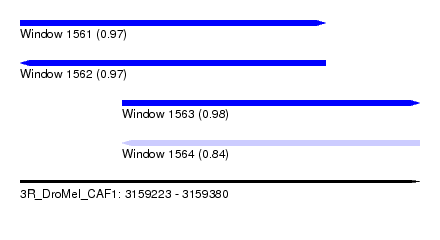
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 3,159,223 – 3,159,380 |
| Length | 157 |
| Max. P | 0.980266 |

| Location | 3,159,223 – 3,159,343 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.81 |
| Mean single sequence MFE | -33.01 |
| Consensus MFE | -31.65 |
| Energy contribution | -31.52 |
| Covariance contribution | -0.13 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.49 |
| Structure conservation index | 0.96 |
| SVM decision value | 1.66 |
| SVM RNA-class probability | 0.970592 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
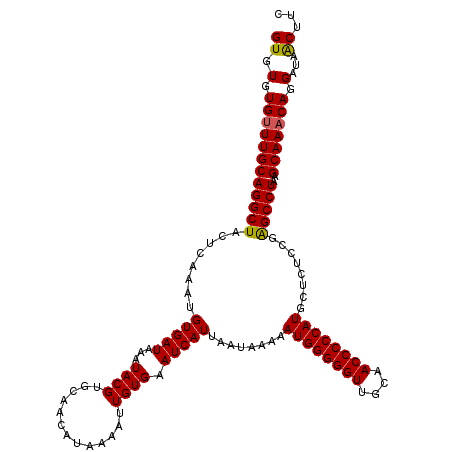
>3R_DroMel_CAF1 3159223 120 + 27905053 GUGUGUGUUUGCAGGCUACUCAAAUGUGAUAAAUACGUGCAACAUAAAAUUGUGAAUCAUUAAUAAAAAUGGGGGUUGCAACCCCCAUUCUCUCUGAGCCUAAGCAAACAGGAUAACUUC ((.(.((((((((((((........(((((...((((.............)))).))))).......(((((((((....))))))))).......)))))..))))))).)...))... ( -32.82) >DroSec_CAF1 7094 120 + 1 GUGUGUGUUUGCAGGCUACUCAAAUGUGAUAAAUACGUGCAACAUAAAAUUGUGAAUCAUUAAUAAAAAUGGGGGUUGCAACCCCCAUGCUCUCCGAGCCUAAGCAAACAGGAUAACUUC ((.(.((((((((((((......(((((.....)))))((..((((....))))..............((((((((....))))))))))......)))))..))))))).)...))... ( -34.10) >DroSim_CAF1 9275 120 + 1 GUGUGUGUUUGCAGGCUACUCAAAUGUGAUAAAUACGUGCAACAUAAAAUUGUGAAUCAUUAAUAAAAAUGGGGGUUGCAACCCCCAUGCUCUCCGAGCCUAAGCAAACAGGAUAACUUC ((.(.((((((((((((......(((((.....)))))((..((((....))))..............((((((((....))))))))))......)))))..))))))).)...))... ( -34.10) >DroYak_CAF1 8636 120 + 1 GUGUGUGCUUGCAGGCUACUCAAAUGUGAUAAAUACGUGCAAAAUAAAAUUGUGAAUCAUUAAUAAAAAUGGGGGUUGCAACCCCCAUUCUCACUGGGCCUAAGCAAACAGGAUAGCUUC .(((.(((((..(((((........((((.........((((.......))))..............(((((((((....))))))))).))))..)))))))))).))).......... ( -31.00) >consensus GUGUGUGUUUGCAGGCUACUCAAAUGUGAUAAAUACGUGCAACAUAAAAUUGUGAAUCAUUAAUAAAAAUGGGGGUUGCAACCCCCAUGCUCUCCGAGCCUAAGCAAACAGGAUAACUUC ((.(.((((((((((((........(((((...((((.............)))).)))))........((((((((....))))))))........)))))..))))))).)...))... (-31.65 = -31.52 + -0.13)

| Location | 3,159,223 – 3,159,343 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.81 |
| Mean single sequence MFE | -33.22 |
| Consensus MFE | -31.66 |
| Energy contribution | -31.98 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.46 |
| Structure conservation index | 0.95 |
| SVM decision value | 1.60 |
| SVM RNA-class probability | 0.966477 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 3159223 120 - 27905053 GAAGUUAUCCUGUUUGCUUAGGCUCAGAGAGAAUGGGGGUUGCAACCCCCAUUUUUAUUAAUGAUUCACAAUUUUAUGUUGCACGUAUUUAUCACAUUUGAGUAGCCUGCAAACACACAC ..........(((((((....(((((((((((((((((((....))))))))))))..................((((.....)))).........))))))).....)))))))..... ( -35.50) >DroSec_CAF1 7094 120 - 1 GAAGUUAUCCUGUUUGCUUAGGCUCGGAGAGCAUGGGGGUUGCAACCCCCAUUUUUAUUAAUGAUUCACAAUUUUAUGUUGCACGUAUUUAUCACAUUUGAGUAGCCUGCAAACACACAC ..........(((((((....((((((((((.((((((((....)))))))).)))..................((((.....)))).........))))))).....)))))))..... ( -30.80) >DroSim_CAF1 9275 120 - 1 GAAGUUAUCCUGUUUGCUUAGGCUCGGAGAGCAUGGGGGUUGCAACCCCCAUUUUUAUUAAUGAUUCACAAUUUUAUGUUGCACGUAUUUAUCACAUUUGAGUAGCCUGCAAACACACAC ..........(((((((....((((((((((.((((((((....)))))))).)))..................((((.....)))).........))))))).....)))))))..... ( -30.80) >DroYak_CAF1 8636 120 - 1 GAAGCUAUCCUGUUUGCUUAGGCCCAGUGAGAAUGGGGGUUGCAACCCCCAUUUUUAUUAAUGAUUCACAAUUUUAUUUUGCACGUAUUUAUCACAUUUGAGUAGCCUGCAAGCACACAC ..........(((((((..((((..(((((((((((((((....)))))))))))))))..........................((((((.......)))))))))))))))))..... ( -35.80) >consensus GAAGUUAUCCUGUUUGCUUAGGCUCAGAGAGAAUGGGGGUUGCAACCCCCAUUUUUAUUAAUGAUUCACAAUUUUAUGUUGCACGUAUUUAUCACAUUUGAGUAGCCUGCAAACACACAC ..........(((((((..((((.....((((((((((((....)))))))))))).............................((((((.......)))))))))))))))))..... (-31.66 = -31.98 + 0.31)

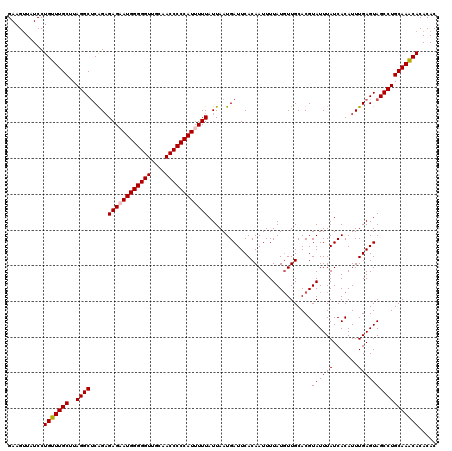
| Location | 3,159,263 – 3,159,380 |
|---|---|
| Length | 117 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.66 |
| Mean single sequence MFE | -32.08 |
| Consensus MFE | -28.90 |
| Energy contribution | -29.15 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.70 |
| Structure conservation index | 0.90 |
| SVM decision value | 1.86 |
| SVM RNA-class probability | 0.980266 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
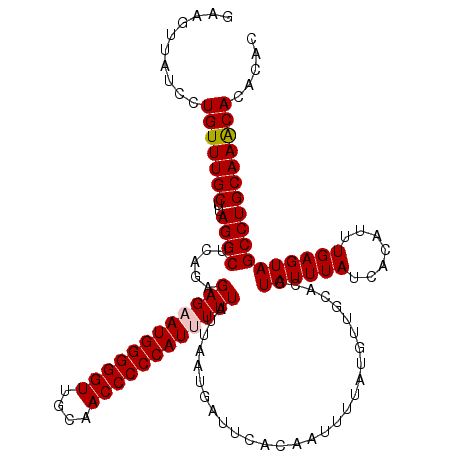
>3R_DroMel_CAF1 3159263 117 + 27905053 AACAUAAAAUUGUGAAUCAUUAAUAAAAAUGGGGGUUGCAACCCCCAUUCUCUCUGAGCCUAAGCAAACAGGAUAACUUCGUCCUGGAAGUCCAUUCCGUUCUGGCGAA---AACAUUUC ..........(((...((.........(((((((((....)))))))))........(((..(((....(((((......)))))((((.....)))))))..))))).---.))).... ( -32.70) >DroSec_CAF1 7134 117 + 1 AACAUAAAAUUGUGAAUCAUUAAUAAAAAUGGGGGUUGCAACCCCCAUGCUCUCCGAGCCUAAGCAAACAGGAUAACUUCGUCCUGGAAGUCCAUUCCGUUCUUGCGAA---AACAUUUC ..........(((...((..........((((((((....))))))))((((...))))....(((((((((((......)))))((((.....))))))..)))))).---.))).... ( -30.70) >DroSim_CAF1 9315 117 + 1 AACAUAAAAUUGUGAAUCAUUAAUAAAAAUGGGGGUUGCAACCCCCAUGCUCUCCGAGCCUAAGCAAACAGGAUAACUUCGUCCUGGAAGUCCAUUCCGUUCUGGCGAA---AACAUUUC ..........(((...((..........((((((((....)))))))).........(((..(((....(((((......)))))((((.....)))))))..))))).---.))).... ( -32.40) >DroYak_CAF1 8676 120 + 1 AAAAUAAAAUUGUGAAUCAUUAAUAAAAAUGGGGGUUGCAACCCCCAUUCUCACUGGGCCUAAGCAAACAGGAUAGCUUCGUCCUGGAAGUCCAUUCCGUUCUGGCGAACGAACCGUUUC ...........((((............(((((((((....))))))))).))))...(((..(((....(((((......)))))((((.....)))))))..)))(((((...))))). ( -32.52) >consensus AACAUAAAAUUGUGAAUCAUUAAUAAAAAUGGGGGUUGCAACCCCCAUGCUCUCCGAGCCUAAGCAAACAGGAUAACUUCGUCCUGGAAGUCCAUUCCGUUCUGGCGAA___AACAUUUC ................((..........((((((((....)))))))).........(((..(((....(((((......)))))((((.....)))))))..)))))............ (-28.90 = -29.15 + 0.25)

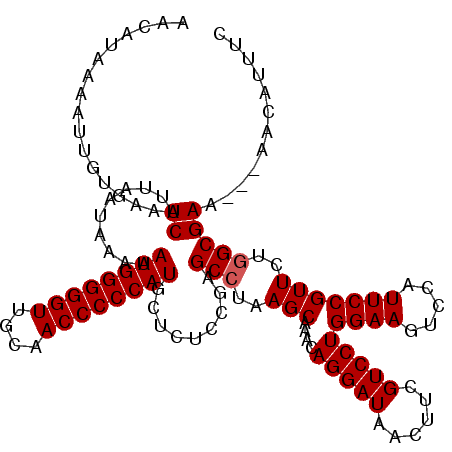
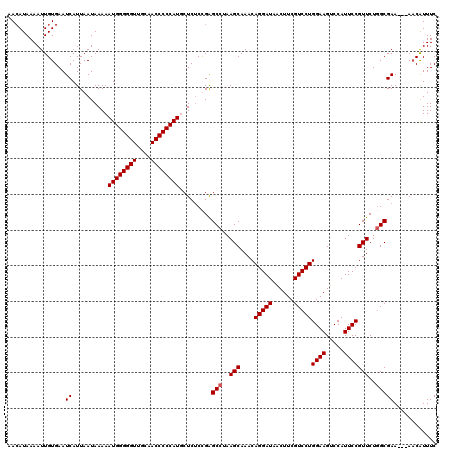
| Location | 3,159,263 – 3,159,380 |
|---|---|
| Length | 117 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.66 |
| Mean single sequence MFE | -35.03 |
| Consensus MFE | -30.29 |
| Energy contribution | -30.85 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.99 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.76 |
| SVM RNA-class probability | 0.844432 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 3159263 117 - 27905053 GAAAUGUU---UUCGCCAGAACGGAAUGGACUUCCAGGACGAAGUUAUCCUGUUUGCUUAGGCUCAGAGAGAAUGGGGGUUGCAACCCCCAUUUUUAUUAAUGAUUCACAAUUUUAUGUU ....(((.---.(((((.(((((((...((((((......)))))).))).)))).....))).....((((((((((((....))))))))))))......))...))).......... ( -34.00) >DroSec_CAF1 7134 117 - 1 GAAAUGUU---UUCGCAAGAACGGAAUGGACUUCCAGGACGAAGUUAUCCUGUUUGCUUAGGCUCGGAGAGCAUGGGGGUUGCAACCCCCAUUUUUAUUAAUGAUUCACAAUUUUAUGUU ((((((((---(((....(((((((...((((((......)))))).))).))))((....))...))))))))((((((....))))))..............)))............. ( -32.40) >DroSim_CAF1 9315 117 - 1 GAAAUGUU---UUCGCCAGAACGGAAUGGACUUCCAGGACGAAGUUAUCCUGUUUGCUUAGGCUCGGAGAGCAUGGGGGUUGCAACCCCCAUUUUUAUUAAUGAUUCACAAUUUUAUGUU ((((((((---((((((.(((((((...((((((......)))))).))).)))).....)))...))))))))((((((....))))))..............)))............. ( -33.90) >DroYak_CAF1 8676 120 - 1 GAAACGGUUCGUUCGCCAGAACGGAAUGGACUUCCAGGACGAAGCUAUCCUGUUUGCUUAGGCCCAGUGAGAAUGGGGGUUGCAACCCCCAUUUUUAUUAAUGAUUCACAAUUUUAUUUU .....(..(((((.(((.((((((.((((.((((......)))))))).)))))).....)))..(((((((((((((((....))))))))))))))))))))..)............. ( -39.80) >consensus GAAAUGUU___UUCGCCAGAACGGAAUGGACUUCCAGGACGAAGUUAUCCUGUUUGCUUAGGCUCAGAGAGAAUGGGGGUUGCAACCCCCAUUUUUAUUAAUGAUUCACAAUUUUAUGUU (((...........(((.((((((.((((.((((......)))))))).)))))).....))).....((((((((((((....))))))))))))........)))............. (-30.29 = -30.85 + 0.56)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:57:06 2006