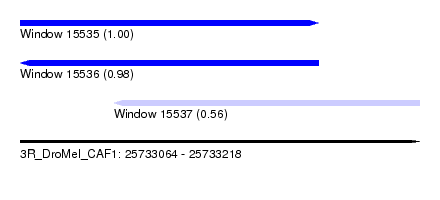
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 25,733,064 – 25,733,218 |
| Length | 154 |
| Max. P | 0.998273 |

| Location | 25,733,064 – 25,733,179 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 74.12 |
| Mean single sequence MFE | -26.96 |
| Consensus MFE | -14.24 |
| Energy contribution | -15.42 |
| Covariance contribution | 1.18 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.67 |
| Structure conservation index | 0.53 |
| SVM decision value | 3.05 |
| SVM RNA-class probability | 0.998273 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 25733064 115 + 27905053 CAACAAUAGAUACAUAUG----UAUGGCAUAAAAUGUAUGCUGCCAGUUGGUUUGUCAUGUUUGCCAUCUGAGCAAGCUAAGAAACUUGUUUAA-ACCUAAUGGCUAAUUAUUGUUAAUU .((((((((((((....)----)))((((((.....))))))((((...((((((....(((((((....).))))))((((...))))..)))-)))...))))...)))))))).... ( -29.70) >DroSec_CAF1 28404 105 + 1 CAACAAUAAAUACACAUG----UAUGGCACGAAAUGUAUGCUGCCACUUGGUU---------UGCCAUCUCAGCAAGCUAAGAAACUUGUUUAA-ACCU-AUGGCUAAUUAUUGUUAAUU .((((((((.((..((((----..(((((((.......)).)))))(((((((---------(((.......))))))))))............-...)-)))..)).)))))))).... ( -25.70) >DroSim_CAF1 28901 109 + 1 CUACAAUAGAUACAUAUGUAUGUAUGGCACAAAACGUAUGCUGCCACUUGGUU---------UGCCAUCUCAGCAAGCUAAGAAACUUGUUUAA-ACCU-AUGGCUAAUUAUUGUUAAUU ..(((((((...(((((((.(((.....)))..)))))))..(((((((((((---------(((.......))))))))))(((....)))..-....-.))))...)))))))..... ( -25.80) >DroEre_CAF1 29660 93 + 1 UAACAAUGGAUAUGUAUG-------------------------GCAGUUUGUUUUCCAUGUUUGCUAUCUAAGCAAGUUAAGAAACUUGUUUAA-ACAU-ACGGCUAAUUAUUGUUAAUU (((((((((...((((((-------------------------((((..((.....))...))))....((((((((((....)))))))))).-.)))-))).....)))))))))... ( -23.30) >DroYak_CAF1 29457 118 + 1 U-ACAAUGGAUAUGUAUAGAUGGAUGGCACACAAUGCAUGUUGCCAGUUGGCUAUCCAUGUUUGCUAUGGAAGCAAGUUAAGAAACUUGUUUAAAACUU-UUGGCUAAUUAUUGUUAAUU .-((((((((((((.((((.....(((((.(((.....)))))))).....)))).)))))).((((.(.(((((((((....)))))))))....)..-.))))....))))))..... ( -30.30) >consensus CAACAAUAGAUACAUAUG____UAUGGCACAAAAUGUAUGCUGCCAGUUGGUU___CAUGUUUGCCAUCUAAGCAAGCUAAGAAACUUGUUUAA_ACCU_AUGGCUAAUUAUUGUUAAUU .((((((((...............(((((............))))).................(((((..(((((((........)))))))........)))))...)))))))).... (-14.24 = -15.42 + 1.18)



| Location | 25,733,064 – 25,733,179 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 74.12 |
| Mean single sequence MFE | -23.04 |
| Consensus MFE | -12.83 |
| Energy contribution | -13.40 |
| Covariance contribution | 0.57 |
| Combinations/Pair | 1.29 |
| Mean z-score | -2.23 |
| Structure conservation index | 0.56 |
| SVM decision value | 1.96 |
| SVM RNA-class probability | 0.983922 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 25733064 115 - 27905053 AAUUAACAAUAAUUAGCCAUUAGGU-UUAAACAAGUUUCUUAGCUUGCUCAGAUGGCAAACAUGACAAACCAACUGGCAGCAUACAUUUUAUGCCAUA----CAUAUGUAUCUAUUGUUG ...((((((((....((((...(((-((...((((((....)))))).....(((.....)))...)))))...)))).(((((.....))))).(((----(....)))).)))))))) ( -24.90) >DroSec_CAF1 28404 105 - 1 AAUUAACAAUAAUUAGCCAU-AGGU-UUAAACAAGUUUCUUAGCUUGCUGAGAUGGCA---------AACCAAGUGGCAGCAUACAUUUCGUGCCAUA----CAUGUGUAUUUAUUGUUG ...(((((((((...(((((-.(((-((...((....((((((....))))))))..)---------))))..)))))...(((((...((((.....----)))))))))))))))))) ( -26.60) >DroSim_CAF1 28901 109 - 1 AAUUAACAAUAAUUAGCCAU-AGGU-UUAAACAAGUUUCUUAGCUUGCUGAGAUGGCA---------AACCAAGUGGCAGCAUACGUUUUGUGCCAUACAUACAUAUGUAUCUAUUGUAG .....((((((....(((((-.(((-((...((....((((((....))))))))..)---------))))..))))).(((((.....))))).((((((....)))))).)))))).. ( -27.10) >DroEre_CAF1 29660 93 - 1 AAUUAACAAUAAUUAGCCGU-AUGU-UUAAACAAGUUUCUUAACUUGCUUAGAUAGCAAACAUGGAAAACAAACUGC-------------------------CAUACAUAUCCAUUGUUA ...(((((((........((-.(((-((((.((((((....)))))).)))))))))....((((...........)-------------------------)))........))))))) ( -15.90) >DroYak_CAF1 29457 118 - 1 AAUUAACAAUAAUUAGCCAA-AAGUUUUAAACAAGUUUCUUAACUUGCUUCCAUAGCAAACAUGGAUAGCCAACUGGCAACAUGCAUUGUGUGCCAUCCAUCUAUACAUAUCCAUUGU-A .....(((((..........-..((((.....(((((....)))))(((.....)))))))((((((.((((....((.....))....)).)).))))))............)))))-. ( -20.70) >consensus AAUUAACAAUAAUUAGCCAU_AGGU_UUAAACAAGUUUCUUAGCUUGCUGAGAUGGCAAACAUG___AACCAACUGGCAGCAUACAUUUUGUGCCAUA____CAUAUGUAUCUAUUGUUG ...((((((((....((((...(((.......(((((....)))))(((.....)))...........)))...)))).(((((.....)))))..................)))))))) (-12.83 = -13.40 + 0.57)



| Location | 25,733,100 – 25,733,218 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 82.42 |
| Mean single sequence MFE | -22.76 |
| Consensus MFE | -15.78 |
| Energy contribution | -15.90 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.61 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.04 |
| SVM RNA-class probability | 0.555585 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 25733100 118 - 27905053 GCUUUACUGUAAGCACACAAAAGUGUUAAAAGCUUGAUUAAUUAACAAUAAUUAGCCAUUAGGU-UUAAACAAGUUUCUUAGCUUGCUCAGAUGGCAAACAUGACAAACCAACUGGCAG ((......((((((((((....))))...(((((((..((((((....))))))(((....)))-.....)))))))....)))))).(((.(((.............))).))))).. ( -24.92) >DroSec_CAF1 28440 108 - 1 GCUUUACUGUAAGCAUAAAAAAGUGUUAAAAGCUUGAUUAAUUAACAAUAAUUAGCCAU-AGGU-UUAAACAAGUUUCUUAGCUUGCUGAGAUGGCA---------AACCAAGUGGCAG (((..(((((((((.((((((..((((.(((.((((..((((((....))))))..)).-)).)-)).))))..))).))))))))).....(((..---------..))))))))).. ( -22.70) >DroSim_CAF1 28941 108 - 1 GCUUUACUGUAAGCAUACAAAAGUGUUAAAAGCUUGAUUAAUUAACAAUAAUUAGCCAU-AGGU-UUAAACAAGUUUCUUAGCUUGCUGAGAUGGCA---------AACCAAGUGGCAG ((((......))))......((((.......))))...((((((....))))))(((((-.(((-((...((....((((((....))))))))..)---------))))..))))).. ( -22.60) >DroEre_CAF1 29678 111 - 1 GCUUU---GUAAGCAUAUCACUACGUUUAAAGCUUGAUUAAUUAACAAUAAUUAGCCGU-AUGU-UUAAACAAGUUUCUUAACUUGCUUAGAUAGCAAACAUGGAAAACAAACUGC--- (((.(---.(((((..........(((((((...((..((((((....))))))..)).-...)-))))))(((((....)))))))))).).)))....................--- ( -19.60) >DroYak_CAF1 29496 115 - 1 GCUUU---CUAAGCAUAGCAAAGUGUUAAAAGCUUGAUUAAUUAACAAUAAUUAGCCAA-AAGUUUUAAACAAGUUUCUUAACUUGCUUCCAUAGCAAACAUGGAUAGCCAACUGGCAA (((..---..(((((.((.((..((((.(((((((..(((((((....)))))))....-))))))).))))..)).)).....)))))....)))...........(((....))).. ( -24.00) >consensus GCUUUACUGUAAGCAUACAAAAGUGUUAAAAGCUUGAUUAAUUAACAAUAAUUAGCCAU_AGGU_UUAAACAAGUUUCUUAGCUUGCUGAGAUGGCAAACAUG___AACCAACUGGCAG ((((......))))..........((((((((((((..((((((....))))))(((....)))......))))))).))))).((((.....))))...................... (-15.78 = -15.90 + 0.12)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:40:45 2006