
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 25,729,175 – 25,729,318 |
| Length | 143 |
| Max. P | 0.588978 |

| Location | 25,729,175 – 25,729,294 |
|---|---|
| Length | 119 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.12 |
| Mean single sequence MFE | -38.08 |
| Consensus MFE | -23.80 |
| Energy contribution | -25.80 |
| Covariance contribution | 2.00 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.61 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.04 |
| SVM RNA-class probability | 0.554489 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 25729175 119 - 27905053 UACCAAAUUCCCC-UUCAAGUCUGCCUCACUUUGGGCAUUGGCUUGACCAGUGGAACCAUCAAUUUGGGGGCCACAAUGGAUGGAAAUUCGGUGGCCGCGCUAUCCAUCAGCACCGCCAA .(((((.((((..-.((((((((((((......)))))..)))))))(((((((..(((......)))...))))..)))..)))).)).)))(((.(.(((.......))).).))).. ( -40.70) >DroPse_CAF1 25970 119 - 1 U-UCAAUUGCUCAAUUGCAGUGUGCCUGACAUUCAAGAUUAGCGUACUAAGUGGGUCAGUAGAUGUAGGUGUCACCAUGGAUGGAAAUUCGAUGACAAAGUUCACCAUCAGCACUGCGUC .-..............(((((((..(((((...((...((((....)))).)).)))))..((((..(((((((...(((((....))))).)))))....))..)))).)))))))... ( -31.10) >DroSec_CAF1 24497 118 - 1 U-UCAAAUUCCCC-UUCAAGUCUGUCUCACUUUGGGCAUUGGCUUGACCAGUGGAACCAUCAAUUUGGGGGCCACAAUGGAUGGAAAUUCGGUGGCCGCGUUCGCCAUCAGCACCACCAA .-.....((((..-.((((((((((((......)))))..)))))))(((((((..(((......)))...))))..)))..))))....(((((..((...........)).))))).. ( -37.10) >DroSim_CAF1 24972 118 - 1 U-UUAAAUUCCCC-UUCAAGUCUGCCUCACUUUGGGCAUUGGCUUGACCAGUGGAACCAUCAAUUUGGGGGCCACAAUGGAUGGAAAUGCGGUGGCCGCGUUCGCCAUCAGCACCACCAA .-...........-.((((((((((((......)))))..)))))))...((((..(((......)))...))))..(((.(((...(((((((((.......)))))).))))))))). ( -41.30) >DroEre_CAF1 25647 117 - 1 U-UAAAAUUCCCC-CU-AAGUCUGCGUAUCCUUGGGCCUCGGCUUGACCAGUGGAACCAUCAAUUUAGGGGCCACAAUGGAUGGGAAUUCGGUGGCCGCGUUCUCCAUCAGCACCACCCA .-...(((((((.-..-..(....)..(((((((((((((((.....)).(((....))).......)))))).))).))))))))))).(((((..((...........)).))))).. ( -36.20) >DroYak_CAF1 25346 118 - 1 U-UCACAUUCUCC-CUCAAGUGUGCCUCACUUUGGGCAUCGGCUUGACCAGUGGAACCAUCAAUUUGGGGGCCACAAUGGAUGGAAAUGCGGUCGCCGCGUUUGCCAUCAGCACCACCAA .-...((((....-.((((((((((((......))))))..))))))...((((..(((......)))...))))))))(((((((((((((...)))))))).)))))........... ( -42.10) >consensus U_UCAAAUUCCCC_UUCAAGUCUGCCUCACUUUGGGCAUUGGCUUGACCAGUGGAACCAUCAAUUUGGGGGCCACAAUGGAUGGAAAUUCGGUGGCCGCGUUCGCCAUCAGCACCACCAA ...............((((((((((((......)))))..)))))))...((((..(((......))).((((((...((((....)))).))))))................))))... (-23.80 = -25.80 + 2.00)

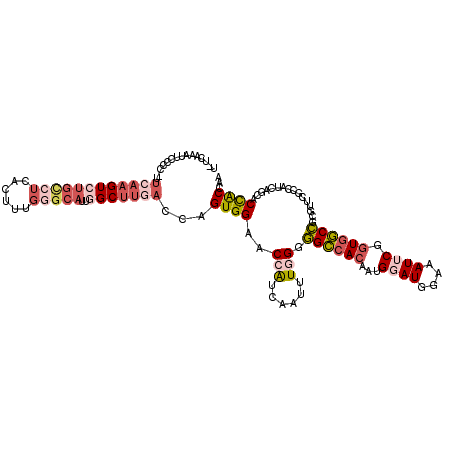

| Location | 25,729,215 – 25,729,318 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 69.49 |
| Mean single sequence MFE | -23.85 |
| Consensus MFE | -13.50 |
| Energy contribution | -15.58 |
| Covariance contribution | 2.09 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.31 |
| Structure conservation index | 0.57 |
| SVM decision value | 0.11 |
| SVM RNA-class probability | 0.588978 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 25729215 103 - 27905053 C-------ACAUUCAUAUAACC-CU----UACCUUA--UACCAAAUUCCCC-UUCAAGUCUGCCUCACUUUGGGCAUUGGCUUGACCAGUGGAACCAUCAAUUUGGGGGCCACAAUGG .-------..............-..----.......--..(((........-.((((((((((((......)))))..)))))))...((((..(((......)))...))))..))) ( -26.30) >DroPse_CAF1 26010 112 - 1 CCAAAACA---GUAGUACAGUACGUUCUAUAAUUCC--U-UCAAUUGCUCAAUUGCAGUGUGCCUGACAUUCAAGAUUAGCGUACUAAGUGGGUCAGUAGAUGUAGGUGUCACCAUGG ........---((..(((..(((((.((((....((--(-.(.(((((......)))))(((((((((......).)))).))))...).)))...))))))))).)))..))..... ( -23.00) >DroSec_CAF1 24537 102 - 1 C-------ACUUUCCUAUAACC-CU----UACCUUA--U-UCAAAUUCCCC-UUCAAGUCUGUCUCACUUUGGGCAUUGGCUUGACCAGUGGAACCAUCAAUUUGGGGGCCACAAUGG .-------............((-(.----.......--.-.....((((.(-(((((((((((((......)))))..)))))))..)).))))(((......))))))......... ( -22.70) >DroSim_CAF1 25012 104 - 1 C-------ACUUUCUUAUAACC-CU----UACCUUACCU-UUAAAUUCCCC-UUCAAGUCUGCCUCACUUUGGGCAUUGGCUUGACCAGUGGAACCAUCAAUUUGGGGGCCACAAUGG .-------............((-(.----..........-.....((((.(-(((((((((((((......)))))..)))))))..)).))))(((......))))))......... ( -25.40) >DroEre_CAF1 25687 93 - 1 U-------AC-------------UUUCCUUACCUCA--U-UAAAAUUCCCC-CU-AAGUCUGCGUAUCCUUGGGCCUCGGCUUGACCAGUGGAACCAUCAAUUUAGGGGCCACAAUGG .-------..-------------.............--.-........(((-((-((((.((.((.(((((((((....))....)))).)))))))...)))))))))......... ( -20.80) >DroYak_CAF1 25386 103 - 1 U-------AU-AACAUAUACCC-CUUCC--CCCUCA--U-UCACAUUCUCC-CUCAAGUGUGCCUCACUUUGGGCAUCGGCUUGACCAGUGGAACCAUCAAUUUGGGGGCCACAAUGG .-------..-...........-.....--......--.-...((((....-.((((((((((((......))))))..))))))...((((..(((......)))...)))))))). ( -24.90) >consensus C_______AC_UUCAUAUAACC_CU____UACCUCA__U_UCAAAUUCCCC_UUCAAGUCUGCCUCACUUUGGGCAUUGGCUUGACCAGUGGAACCAUCAAUUUGGGGGCCACAAUGG .....................................................((((((((((((......)))))..)))))))...((((..((.........))..))))..... (-13.50 = -15.58 + 2.09)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:40:42 2006