
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 25,706,694 – 25,706,858 |
| Length | 164 |
| Max. P | 0.999312 |

| Location | 25,706,694 – 25,706,797 |
|---|---|
| Length | 103 |
| Sequences | 5 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 84.52 |
| Mean single sequence MFE | -23.02 |
| Consensus MFE | -13.82 |
| Energy contribution | -14.94 |
| Covariance contribution | 1.12 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.18 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.18 |
| SVM RNA-class probability | 0.620153 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 25706694 103 + 27905053 AAUAACUCAAAAAAUGGCACAUUAUAUGAGGGAAUUUCUAUUGUUCAU---------UAAAUGUGUGUAUAAUGCGGGAAAAAAUUUGAAUUUUUCCAUGCAUUACACAAGU ................(((((((..(((((.((.......)).)))))---------..)))))))((.((((((((((((((.......))))))).))))))).)).... ( -25.30) >DroSec_CAF1 11314 103 + 1 AUUGACUCCAAAAAUGGCGCAUUAUAUGAGGGAAUUUCUAUUAUUCAU---------UAAACGUGUGUAUAAUGCGCGGAAAAAUUUUAAUUUUUCCAUGCAUUACACAAGU ............((((((((((((((((.(.((((.......))))..---------....)...))))))))))))((((((((....))))))))...))))........ ( -24.00) >DroSim_CAF1 11523 112 + 1 AUUAACUCCAAAAAUGGCGCAUUAUAUGAGGGAAUUUCUAUUAUUCAUUAAAUUCAUUAAAUGUGUGUAUAAUGCGCGGAAAAAUUUUAGUUUUUCCAUGCAUUACACAAGU ................(((((((..(((((.((((.......))))......)))))..)))))))((.(((((((.((((((((....)))))))).))))))).)).... ( -27.50) >DroEre_CAF1 12567 102 + 1 GUUAGCUCCAA-GAUGGCACUUUAUAUGAGGGAAUUUCUAUUAUUCAC---------UAAAUGUGGAUAUAAUGCGCAGAAAAAUUGCAAUCUCUCCAUGCAUUACACAAGC ....((.((..-...))..........((((((........(((((((---------.....)))))))......((((.....))))..))))))...))........... ( -17.70) >DroYak_CAF1 11722 103 + 1 GUUAACUCCAAAAAUGCCACAUUAUACGAGGGAAUUUCUAUUAUUCAC---------UAAGUGUGGCUAUAAUGCGGUUAAAAAUUUCAAUUUUUCCAUGCAUUACACAAGC ...............((((((((.....((.((((.......)))).)---------).))))))))..(((((((...((((((....))))))...)))))))....... ( -20.60) >consensus AUUAACUCCAAAAAUGGCACAUUAUAUGAGGGAAUUUCUAUUAUUCAU_________UAAAUGUGUGUAUAAUGCGCGGAAAAAUUUCAAUUUUUCCAUGCAUUACACAAGU .....(((....((((...))))....))).((((.......))))...............((((....(((((((.((((((((....)))))))).)))))))))))... (-13.82 = -14.94 + 1.12)



| Location | 25,706,694 – 25,706,797 |
|---|---|
| Length | 103 |
| Sequences | 5 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 84.52 |
| Mean single sequence MFE | -23.42 |
| Consensus MFE | -17.24 |
| Energy contribution | -16.96 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.33 |
| Mean z-score | -2.51 |
| Structure conservation index | 0.74 |
| SVM decision value | 1.52 |
| SVM RNA-class probability | 0.961118 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 25706694 103 - 27905053 ACUUGUGUAAUGCAUGGAAAAAUUCAAAUUUUUUCCCGCAUUAUACACACAUUUA---------AUGAACAAUAGAAAUUCCCUCAUAUAAUGUGCCAUUUUUUGAGUUAUU (((((((((((((..(((((((.......))))))).))))))))).((((((..---------((((.....(....)....))))..)))))).........)))).... ( -24.30) >DroSec_CAF1 11314 103 - 1 ACUUGUGUAAUGCAUGGAAAAAUUAAAAUUUUUCCGCGCAUUAUACACACGUUUA---------AUGAAUAAUAGAAAUUCCCUCAUAUAAUGCGCCAUUUUUGGAGUCAAU ...((((((((((.(((((((((....))))))))).))))))))))(.((((..---------((((.....(....)....))))..)))).)(((....)))....... ( -24.90) >DroSim_CAF1 11523 112 - 1 ACUUGUGUAAUGCAUGGAAAAACUAAAAUUUUUCCGCGCAUUAUACACACAUUUAAUGAAUUUAAUGAAUAAUAGAAAUUCCCUCAUAUAAUGCGCCAUUUUUGGAGUUAAU ...((((((((((.((((((((......)))))))).))))))))))..........((((((............)))))).......((((...(((....))).)))).. ( -24.80) >DroEre_CAF1 12567 102 - 1 GCUUGUGUAAUGCAUGGAGAGAUUGCAAUUUUUCUGCGCAUUAUAUCCACAUUUA---------GUGAAUAAUAGAAAUUCCCUCAUAUAAAGUGCCAUC-UUGGAGCUAAC (((((((((((((..((((((((....))))))))..))))))))).(((.((((---------((((.....(....)....)))).))))))).....-...)))).... ( -22.70) >DroYak_CAF1 11722 103 - 1 GCUUGUGUAAUGCAUGGAAAAAUUGAAAUUUUUAACCGCAUUAUAGCCACACUUA---------GUGAAUAAUAGAAAUUCCCUCGUAUAAUGUGGCAUUUUUGGAGUUAAC ((((.((((((((....((((((....))))))....))))))))((((((...(---------(.((((.......)))).)).......)))))).......)))).... ( -20.40) >consensus ACUUGUGUAAUGCAUGGAAAAAUUAAAAUUUUUCCGCGCAUUAUACACACAUUUA_________AUGAAUAAUAGAAAUUCCCUCAUAUAAUGUGCCAUUUUUGGAGUUAAU (((((((((((((.(((((((((....))))))))).)))))))))..................((((.....(....)....)))).................)))).... (-17.24 = -16.96 + -0.28)

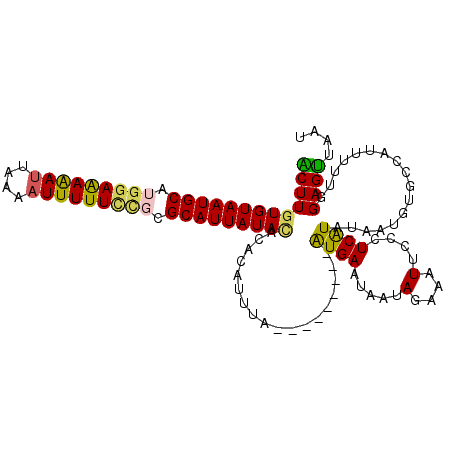
| Location | 25,706,726 – 25,706,837 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 88.39 |
| Mean single sequence MFE | -23.76 |
| Consensus MFE | -18.36 |
| Energy contribution | -20.24 |
| Covariance contribution | 1.88 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.18 |
| Structure conservation index | 0.77 |
| SVM decision value | 1.21 |
| SVM RNA-class probability | 0.930483 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 25706726 111 + 27905053 AAUUUCUAUUGUUCAU---------UAAAUGUGUGUAUAAUGCGGGAAAAAAUUUGAAUUUUUCCAUGCAUUACACAAGUGCAUUUCAAAAGCGAAACGUAUAUAAUUGAAAUAACUCGG .(((((.(((((....---------......((((((..((((((((((((.......))))))).))))))))))).((((.((((......)))).))))))))).)))))....... ( -25.50) >DroSec_CAF1 11346 111 + 1 AAUUUCUAUUAUUCAU---------UAAACGUGUGUAUAAUGCGCGGAAAAAUUUUAAUUUUUCCAUGCAUUACACAAGUGCAUUUCAAAAGCGAAUCGUAUAUAAUUGAAAUAACUCGG .(((((.(((((....---------......((((((..(((((.((((((((....)))))))).))))))))))).((((..(((......)))..))))))))).)))))....... ( -26.40) >DroSim_CAF1 11555 120 + 1 AAUUUCUAUUAUUCAUUAAAUUCAUUAAAUGUGUGUAUAAUGCGCGGAAAAAUUUUAGUUUUUCCAUGCAUUACACAAGUGCAUUUCAAAAGCGAAUCGUAUAUAAUUGAAAUAACUCGG .(((((.(((((...................((((((..(((((.((((((((....)))))))).))))))))))).((((..(((......)))..))))))))).)))))....... ( -27.00) >DroEre_CAF1 12598 111 + 1 AAUUUCUAUUAUUCAC---------UAAAUGUGGAUAUAAUGCGCAGAAAAAUUGCAAUCUCUCCAUGCAUUACACAAGCGCAUUUCAAUAGCGAAACGUAUAUAAUUGAAAUAACUCGG .(((((...(((((((---------.....))))))).(((((((........((((.........))))........))))))).......................)))))....... ( -20.19) >DroYak_CAF1 11754 111 + 1 AAUUUCUAUUAUUCAC---------UAAGUGUGGCUAUAAUGCGGUUAAAAAUUUCAAUUUUUCCAUGCAUUACACAAGCGCAUUUCAAUAGCGAAACGUAUAUAAUUGAAAUAGCUCGG .(((((.(((((..((---------...((((.....(((((((...((((((....))))))...))))))).....)))).((((......)))).))..))))).)))))....... ( -19.70) >consensus AAUUUCUAUUAUUCAU_________UAAAUGUGUGUAUAAUGCGCGGAAAAAUUUCAAUUUUUCCAUGCAUUACACAAGUGCAUUUCAAAAGCGAAACGUAUAUAAUUGAAAUAACUCGG .(((((.(((((.................((((....(((((((.((((((((....)))))))).))))))))))).((((.((((......)))).))))))))).)))))....... (-18.36 = -20.24 + 1.88)



| Location | 25,706,726 – 25,706,837 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.39 |
| Mean single sequence MFE | -25.56 |
| Consensus MFE | -22.77 |
| Energy contribution | -22.89 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.23 |
| Mean z-score | -2.67 |
| Structure conservation index | 0.89 |
| SVM decision value | 3.50 |
| SVM RNA-class probability | 0.999312 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 25706726 111 - 27905053 CCGAGUUAUUUCAAUUAUAUACGUUUCGCUUUUGAAAUGCACUUGUGUAAUGCAUGGAAAAAUUCAAAUUUUUUCCCGCAUUAUACACACAUUUA---------AUGAACAAUAGAAAUU ..((((((((((((.................)))))))).))))(((((((((..(((((((.......))))))).))))))))).........---------................ ( -24.33) >DroSec_CAF1 11346 111 - 1 CCGAGUUAUUUCAAUUAUAUACGAUUCGCUUUUGAAAUGCACUUGUGUAAUGCAUGGAAAAAUUAAAAUUUUUCCGCGCAUUAUACACACGUUUA---------AUGAAUAAUAGAAAUU ....(((((((..((((...(((....((.........))...((((((((((.(((((((((....))))))))).))))))))))..))).))---------)))))))))....... ( -27.90) >DroSim_CAF1 11555 120 - 1 CCGAGUUAUUUCAAUUAUAUACGAUUCGCUUUUGAAAUGCACUUGUGUAAUGCAUGGAAAAACUAAAAUUUUUCCGCGCAUUAUACACACAUUUAAUGAAUUUAAUGAAUAAUAGAAAUU .......(((((.(((((.((.(((((((.........))...((((((((((.((((((((......)))))))).))))))))))..........)))))...)).))))).))))). ( -27.40) >DroEre_CAF1 12598 111 - 1 CCGAGUUAUUUCAAUUAUAUACGUUUCGCUAUUGAAAUGCGCUUGUGUAAUGCAUGGAGAGAUUGCAAUUUUUCUGCGCAUUAUAUCCACAUUUA---------GUGAAUAAUAGAAAUU .......(((((.(((((...(((((((....)))))))((((((((((((((..((((((((....))))))))..))))))....))))...)---------))).))))).))))). ( -25.70) >DroYak_CAF1 11754 111 - 1 CCGAGCUAUUUCAAUUAUAUACGUUUCGCUAUUGAAAUGCGCUUGUGUAAUGCAUGGAAAAAUUGAAAUUUUUAACCGCAUUAUAGCCACACUUA---------GUGAAUAAUAGAAAUU .((((((((((((((...............))))))))).)))))((((((((....((((((....))))))....))))))))..(((.....---------)))............. ( -22.46) >consensus CCGAGUUAUUUCAAUUAUAUACGUUUCGCUUUUGAAAUGCACUUGUGUAAUGCAUGGAAAAAUUAAAAUUUUUCCGCGCAUUAUACACACAUUUA_________AUGAAUAAUAGAAAUU ..((((((((((((.................)))))))).))))(((((((((.(((((((((....))))))))).))))))))).................................. (-22.77 = -22.89 + 0.12)



| Location | 25,706,757 – 25,706,858 |
|---|---|
| Length | 101 |
| Sequences | 5 |
| Columns | 101 |
| Reading direction | reverse |
| Mean pairwise identity | 87.23 |
| Mean single sequence MFE | -26.18 |
| Consensus MFE | -19.07 |
| Energy contribution | -19.11 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.23 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.86 |
| SVM RNA-class probability | 0.869483 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 25706757 101 - 27905053 GCAUGAAGAGAGUGAGUCGCACCGAGUUAUUUCAAUUAUAUACGUUUCGCUUUUGAAAUGCACUUGUGUAAUGCAUGGAAAAAUUCAAAUUUUUUCCCGCA ((((...(((((((((.((.....((((.....)))).....)).)))))))))...))))..........(((..(((((((.......))))))).))) ( -23.70) >DroSec_CAF1 11377 101 - 1 GCAUGAAGAGAGCGAGUCGCACCGAGUUAUUUCAAUUAUAUACGAUUCGCUUUUGAAAUGCACUUGUGUAAUGCAUGGAAAAAUUAAAAUUUUUCCGCGCA ((((...((((((((((((.....((((.....)))).....))))))))))))...))))..........(((.(((((((((....))))))))).))) ( -33.60) >DroSim_CAF1 11595 101 - 1 GCAUGAAGAGAGCGAGUCGCACCGAGUUAUUUCAAUUAUAUACGAUUCGCUUUUGAAAUGCACUUGUGUAAUGCAUGGAAAAACUAAAAUUUUUCCGCGCA ((((...((((((((((((.....((((.....)))).....))))))))))))...))))..........(((.((((((((......)))))))).))) ( -32.50) >DroEre_CAF1 12629 87 - 1 --------------AGUCGCACCGAGUUAUUUCAAUUAUAUACGUUUCGCUAUUGAAAUGCGCUUGUGUAAUGCAUGGAGAGAUUGCAAUUUUUCUGCGCA --------------.((..(((.(((((((((((((...............))))))))).)))))))..))((..((((((((....))))))))..)). ( -20.56) >DroYak_CAF1 11785 101 - 1 GCAUGAAGAGUGUGAGUCGCACCGAGCUAUUUCAAUUAUAUACGUUUCGCUAUUGAAAUGCGCUUGUGUAAUGCAUGGAAAAAUUGAAAUUUUUAACCGCA ((.........(((.((..(((.(((((((((((((...............))))))))).)))))))..)).)))((((((((....))))))..)))). ( -20.56) >consensus GCAUGAAGAGAGCGAGUCGCACCGAGUUAUUUCAAUUAUAUACGUUUCGCUUUUGAAAUGCACUUGUGUAAUGCAUGGAAAAAUUAAAAUUUUUCCGCGCA ...............((..(((.((((((((((((.................)))))))).)))))))..))((.(((((((((....))))))))).)). (-19.07 = -19.11 + 0.04)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:40:36 2006