
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 25,691,570 – 25,691,704 |
| Length | 134 |
| Max. P | 0.995337 |

| Location | 25,691,570 – 25,691,668 |
|---|---|
| Length | 98 |
| Sequences | 5 |
| Columns | 98 |
| Reading direction | forward |
| Mean pairwise identity | 90.70 |
| Mean single sequence MFE | -21.31 |
| Consensus MFE | -18.12 |
| Energy contribution | -18.04 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.85 |
| SVM decision value | 1.46 |
| SVM RNA-class probability | 0.955689 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 25691570 98 + 27905053 GUAGACUUUAUCUAAAAUGCAGGUACUUAAAUGAUAAAUAUAUAUUUGCCAAUAUGGCUUGUUCGAGUUCGAGCAAUAAUUAAUUGGCUUUUUCCAUA (((...((((((....................))))))..)))....((((((.....(((((((....)))))))......)))))).......... ( -16.55) >DroSec_CAF1 7374 98 + 1 CUAGACUUUAUCUAAAAUGCAGGUACUUAAAUGAUAAAUAUAUCUUUGCCAAUUUGGCUUGCUCGAGUUCGAGCAGUAAUUAAUUGGCUUUUUCCAUA .((((.....)))).......((.........((((....))))...(((((((..(((.(((((....))))))))....))))))).....))... ( -22.70) >DroSim_CAF1 7388 98 + 1 GUAGACUUUAUCUAGAAUGCAGGUACUUAAAUGAUAAAUAUAUCUUUGCCAAUUUGGCUUGCUCGAGUUCGAGCAGUAAUUAAUUGGCUUUUUCCAUA (((..((......))..))).((.........((((....))))...(((((((..(((.(((((....))))))))....))))))).....))... ( -23.80) >DroEre_CAF1 7898 96 + 1 GUGGACUUUAUCUAAAAUGCAGGUACUUAAAUGAUUA--AUAUCUUUGCCAAUUUCGAUUGUUCGAGUUCGAGCAAUAAUUAAUUGGCUUUUUCUAUA (((((...............(((((.((((....)))--))))))..(((((((...((((((((....))))))))....)))))))....))))). ( -23.10) >DroYak_CAF1 7891 96 + 1 AUGGGCUUUAUCGAAAAUGCAGGUACUUAAAUGAUUU--AUAUCUUUGCCAAUUUUGUUUGCUCGCGUUCGAGCAAUAAUUAAUUGGCUUUUUCUAUA ............(((((...............(((..--..)))...(((((((....(((((((....))))))).....))))))).))))).... ( -20.40) >consensus GUAGACUUUAUCUAAAAUGCAGGUACUUAAAUGAUAAAUAUAUCUUUGCCAAUUUGGCUUGCUCGAGUUCGAGCAAUAAUUAAUUGGCUUUUUCCAUA ........(((((.......)))))......................(((((((....(((((((....))))))).....))))))).......... (-18.12 = -18.04 + -0.08)

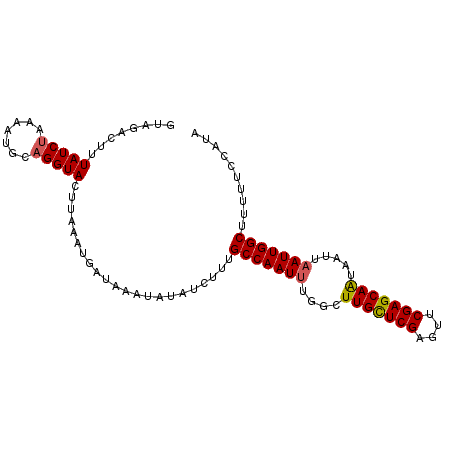

| Location | 25,691,570 – 25,691,668 |
|---|---|
| Length | 98 |
| Sequences | 5 |
| Columns | 98 |
| Reading direction | reverse |
| Mean pairwise identity | 90.70 |
| Mean single sequence MFE | -20.06 |
| Consensus MFE | -16.14 |
| Energy contribution | -17.54 |
| Covariance contribution | 1.40 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.50 |
| Structure conservation index | 0.80 |
| SVM decision value | 2.16 |
| SVM RNA-class probability | 0.989330 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 25691570 98 - 27905053 UAUGGAAAAAGCCAAUUAAUUAUUGCUCGAACUCGAACAAGCCAUAUUGGCAAAUAUAUAUUUAUCAUUUAAGUACCUGCAUUUUAGAUAAAGUCUAC ...((.....((((((......(((.(((....))).))).....)))))).......((((((.....)))))))).......(((((...))))). ( -17.20) >DroSec_CAF1 7374 98 - 1 UAUGGAAAAAGCCAAUUAAUUACUGCUCGAACUCGAGCAAGCCAAAUUGGCAAAGAUAUAUUUAUCAUUUAAGUACCUGCAUUUUAGAUAAAGUCUAG ...((.....(((((((......((((((....)))))).....)))))))...((((....)))).........)).......(((((...))))). ( -23.20) >DroSim_CAF1 7388 98 - 1 UAUGGAAAAAGCCAAUUAAUUACUGCUCGAACUCGAGCAAGCCAAAUUGGCAAAGAUAUAUUUAUCAUUUAAGUACCUGCAUUCUAGAUAAAGUCUAC ..(((((...(((((((......((((((....)))))).....)))))))...((((....))))...............)))))............ ( -23.20) >DroEre_CAF1 7898 96 - 1 UAUAGAAAAAGCCAAUUAAUUAUUGCUCGAACUCGAACAAUCGAAAUUGGCAAAGAUAU--UAAUCAUUUAAGUACCUGCAUUUUAGAUAAAGUCCAC ..........(((((((....((((.(((....))).))))...)))))))...(((..--..)))(((((((.........)))))))......... ( -15.10) >DroYak_CAF1 7891 96 - 1 UAUAGAAAAAGCCAAUUAAUUAUUGCUCGAACGCGAGCAAACAAAAUUGGCAAAGAUAU--AAAUCAUUUAAGUACCUGCAUUUUCGAUAAAGCCCAU ....(((((.(((((((.....(((((((....)))))))....)))))))...(((..--..)))...............)))))............ ( -21.60) >consensus UAUGGAAAAAGCCAAUUAAUUAUUGCUCGAACUCGAGCAAGCCAAAUUGGCAAAGAUAUAUUUAUCAUUUAAGUACCUGCAUUUUAGAUAAAGUCUAC ..(((((...(((((((.....(((((((....)))))))....)))))))...((((....))))...............)))))............ (-16.14 = -17.54 + 1.40)



| Location | 25,691,600 – 25,691,704 |
|---|---|
| Length | 104 |
| Sequences | 5 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 88.58 |
| Mean single sequence MFE | -24.48 |
| Consensus MFE | -18.88 |
| Energy contribution | -19.00 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.30 |
| Structure conservation index | 0.77 |
| SVM decision value | 1.37 |
| SVM RNA-class probability | 0.947406 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 25691600 104 + 27905053 AUGAUAAAUAUAUAUUUGCCAAUAUGGCUUGUUCGAGUUCGAGCAAUAAUUAAUUGGCUUUUUCCAUAAUUAAUAAGCAAUUUAGAUGCAGUUA----GUUAGUUUGA ....(((((........((((((.....(((((((....)))))))......))))))........((((((((..(((.......))).))))----))))))))). ( -20.70) >DroSec_CAF1 7404 108 + 1 AUGAUAAAUAUAUCUUUGCCAAUUUGGCUUGCUCGAGUUCGAGCAGUAAUUAAUUGGCUUUUUCCAUAAUUUAUAAGCAAUUAAGAUGAAGUCAUGUGCUUAGUUUGA ..((((....))))...(((((((..(((.(((((....))))))))....)))))))...............((((((.....(((...)))...))))))...... ( -25.80) >DroSim_CAF1 7418 107 + 1 AUGAUAAAUAUAUCUUUGCCAAUUUGGCUUGCUCGAGUUCGAGCAGUAAUUAAUUGGCUUUUUCCAUAAUUUAUAAGCAAUUAAGAUGAAGUCAUG-GCUUAGUUUGA ....(((((.(((((..(((((((..(((.(((((....))))))))....)))))))...............(((....))))))))((((....-)))).))))). ( -25.20) >DroEre_CAF1 7928 106 + 1 AUGAUUA--AUAUCUUUGCCAAUUUCGAUUGUUCGAGUUCGAGCAAUAAUUAAUUGGCUUUUUCUAUAAUUUAUAAGCAAUUUAGAUGUAGUUAUAUGCUUAGUUUGA ..(((((--........(((((((...((((((((....))))))))....)))))))........)))))..((((((...(((......)))..))))))...... ( -23.49) >DroYak_CAF1 7921 106 + 1 AUGAUUU--AUAUCUUUGCCAAUUUUGUUUGCUCGCGUUCGAGCAAUAAUUAAUUGGCUUUUUCUAUAAUUCAUAAGCAAUUUAGACGCAGUUAUAUGCUUAGUUUGA ((((.((--(((.....(((((((....(((((((....))))))).....)))))))......))))).)))).......(((((((((......)))...)))))) ( -27.20) >consensus AUGAUAAAUAUAUCUUUGCCAAUUUGGCUUGCUCGAGUUCGAGCAAUAAUUAAUUGGCUUUUUCCAUAAUUUAUAAGCAAUUUAGAUGAAGUUAUAUGCUUAGUUUGA .................(((((((....(((((((....))))))).....)))))))...............((((((.....(((...)))...))))))...... (-18.88 = -19.00 + 0.12)



| Location | 25,691,600 – 25,691,704 |
|---|---|
| Length | 104 |
| Sequences | 5 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 88.58 |
| Mean single sequence MFE | -21.14 |
| Consensus MFE | -16.41 |
| Energy contribution | -18.21 |
| Covariance contribution | 1.80 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.20 |
| Structure conservation index | 0.78 |
| SVM decision value | 2.57 |
| SVM RNA-class probability | 0.995337 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 25691600 104 - 27905053 UCAAACUAAC----UAACUGCAUCUAAAUUGCUUAUUAAUUAUGGAAAAAGCCAAUUAAUUAUUGCUCGAACUCGAACAAGCCAUAUUGGCAAAUAUAUAUUUAUCAU ..........----.....(((.......)))....(((.((((......((((((......(((.(((....))).))).....)))))).....)))).))).... ( -15.20) >DroSec_CAF1 7404 108 - 1 UCAAACUAAGCACAUGACUUCAUCUUAAUUGCUUAUAAAUUAUGGAAAAAGCCAAUUAAUUACUGCUCGAACUCGAGCAAGCCAAAUUGGCAAAGAUAUAUUUAUCAU ......((((((.(((....)))......))))))...............(((((((......((((((....)))))).....)))))))...((((....)))).. ( -23.80) >DroSim_CAF1 7418 107 - 1 UCAAACUAAGC-CAUGACUUCAUCUUAAUUGCUUAUAAAUUAUGGAAAAAGCCAAUUAAUUACUGCUCGAACUCGAGCAAGCCAAAUUGGCAAAGAUAUAUUUAUCAU ..........(-(((((......................)))))).....(((((((......((((((....)))))).....)))))))...((((....)))).. ( -23.05) >DroEre_CAF1 7928 106 - 1 UCAAACUAAGCAUAUAACUACAUCUAAAUUGCUUAUAAAUUAUAGAAAAAGCCAAUUAAUUAUUGCUCGAACUCGAACAAUCGAAAUUGGCAAAGAUAU--UAAUCAU ......((((((.................))))))...............(((((((....((((.(((....))).))))...)))))))...(((..--..))).. ( -18.13) >DroYak_CAF1 7921 106 - 1 UCAAACUAAGCAUAUAACUGCGUCUAAAUUGCUUAUGAAUUAUAGAAAAAGCCAAUUAAUUAUUGCUCGAACGCGAGCAAACAAAAUUGGCAAAGAUAU--AAAUCAU ......((((((.................))))))(((.(((((......(((((((.....(((((((....)))))))....))))))).....)))--)).))). ( -25.53) >consensus UCAAACUAAGCACAUAACUGCAUCUAAAUUGCUUAUAAAUUAUGGAAAAAGCCAAUUAAUUAUUGCUCGAACUCGAGCAAGCCAAAUUGGCAAAGAUAUAUUUAUCAU ......((((((.................))))))...............(((((((.....(((((((....)))))))....)))))))................. (-16.41 = -18.21 + 1.80)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:40:29 2006