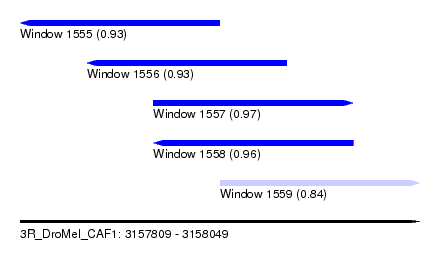
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 3,157,809 – 3,158,049 |
| Length | 240 |
| Max. P | 0.974026 |

| Location | 3,157,809 – 3,157,929 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.81 |
| Mean single sequence MFE | -36.45 |
| Consensus MFE | -34.31 |
| Energy contribution | -34.88 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.94 |
| SVM decision value | 1.21 |
| SVM RNA-class probability | 0.931417 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 3157809 120 - 27905053 CUCAGACUUCAAUCAAGUUUGGGGGCCUUUGAUCAGCUGCCGUUGGCAUCAAUCGAAGACCCCCGAUCCUUGAACCUAUUUGAUCUGUGCUUACACUGGUAAAAACUGAGAUACCGCUGA (((((.......(((((.(((((((.(((((((..(.((((...)))).).))))))).)))))))..))))).....(((.(((.(((....))).))).))).))))).......... ( -39.20) >DroSec_CAF1 5663 120 - 1 CUCAGACUUCAAUCAAGUUUGGGGGCCUUUGAUCAGCUGCCGUUGGCAUCAAUCGAAGACUCCCGAUCCUUGAACCUAUGUGAUCUGUGCUUACACUGGUAAAAAAUGAGACACCGCCGA ((((........(((((.(((((((.(((((((..(.((((...)))).).))))))).)))))))..)))))(((..(((((.......)))))..)))......)))).......... ( -36.20) >DroSim_CAF1 7837 120 - 1 CUCAGACUUCAAUCAAGUUUGGGGGCCUUUGAUCAGCUGCCGUUGGCAUCAAUCGAAGACUCCCGAUCCUUGAACCUAUGUGAUCUGUGCCUACACUGGUAAAAACCAAGAUACCGUUAA ............(((((.(((((((.(((((((..(.((((...)))).).))))))).)))))))..)))))....(((..(((((((....)))((((....))))))))..)))... ( -33.50) >DroYak_CAF1 5367 120 - 1 CUCGGACUUCAAUCAAGUUUGGGGGCCUUUGAUCAGCUGCCGUUGGCAUCAAUCGAGGACCACCGAUUCUUGAGCCUAUGUGAUCUGUGUUUGCACUGGUGAAAACUGAGAUACAGUUGA (((((..(((..(((((..(.((((.(((((((..(.((((...)))).).))))))).)).)).)..)))))(((..((..(.......)..))..))))))..))))).......... ( -36.90) >consensus CUCAGACUUCAAUCAAGUUUGGGGGCCUUUGAUCAGCUGCCGUUGGCAUCAAUCGAAGACCCCCGAUCCUUGAACCUAUGUGAUCUGUGCUUACACUGGUAAAAACUGAGAUACCGCUGA (((((.......(((((.(((((((.(((((((..(.((((...)))).).))))))).)))))))..)))))(((..(((((.......)))))..))).....))))).......... (-34.31 = -34.88 + 0.56)

| Location | 3,157,849 – 3,157,969 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.11 |
| Mean single sequence MFE | -33.23 |
| Consensus MFE | -30.59 |
| Energy contribution | -30.90 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.16 |
| Structure conservation index | 0.92 |
| SVM decision value | 1.17 |
| SVM RNA-class probability | 0.925039 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 3157849 120 - 27905053 AUCUAAUAAAUUAUCAUUUGCACUGCAAUCGUCAGUGUCCCUCAGACUUCAAUCAAGUUUGGGGGCCUUUGAUCAGCUGCCGUUGGCAUCAAUCGAAGACCCCCGAUCCUUGAACCUAUU ...................(((((((....).))))))..............(((((.(((((((.(((((((..(.((((...)))).).))))))).)))))))..)))))....... ( -35.70) >DroSec_CAF1 5703 120 - 1 AUCUAAUAAAUUAUCAUUUGCACUGCAAUCGUCAGUGUCCCUCAGACUUCAAUCAAGUUUGGGGGCCUUUGAUCAGCUGCCGUUGGCAUCAAUCGAAGACUCCCGAUCCUUGAACCUAUG ...................(((((((....).))))))..............(((((.(((((((.(((((((..(.((((...)))).).))))))).)))))))..)))))....... ( -32.70) >DroSim_CAF1 7877 120 - 1 AUCUAAUAAAUUAUCAUUUGCACUGCAAUCGUCAGUGUCCCUCAGACUUCAAUCAAGUUUGGGGGCCUUUGAUCAGCUGCCGUUGGCAUCAAUCGAAGACUCCCGAUCCUUGAACCUAUG ...................(((((((....).))))))..............(((((.(((((((.(((((((..(.((((...)))).).))))))).)))))))..)))))....... ( -32.70) >DroYak_CAF1 5407 120 - 1 AUCUAAUAAAUUAUCAUUUGAACUGCAAUCGUCAGCGUCCCUCGGACUUCAAUCAAGUUUGGGGGCCUUUGAUCAGCUGCCGUUGGCAUCAAUCGAGGACCACCGAUUCUUGAGCCUAUG ....................(((.(((.(.(((((.(.(((((((((((.....))))))))))).).))))).)..))).)))(((.(((((((.(.....)))))...)))))).... ( -31.80) >consensus AUCUAAUAAAUUAUCAUUUGCACUGCAAUCGUCAGUGUCCCUCAGACUUCAAUCAAGUUUGGGGGCCUUUGAUCAGCUGCCGUUGGCAUCAAUCGAAGACCCCCGAUCCUUGAACCUAUG ...................(((((((....).))))))..............(((((.(((((((.(((((((..(.((((...)))).).))))))).)))))))..)))))....... (-30.59 = -30.90 + 0.31)

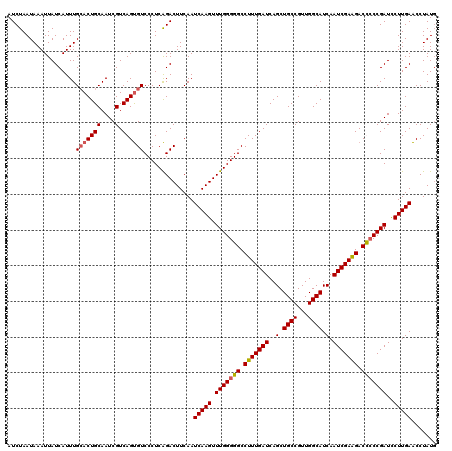
| Location | 3,157,889 – 3,158,009 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.42 |
| Mean single sequence MFE | -39.70 |
| Consensus MFE | -35.67 |
| Energy contribution | -36.30 |
| Covariance contribution | 0.63 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.79 |
| Structure conservation index | 0.90 |
| SVM decision value | 1.72 |
| SVM RNA-class probability | 0.974026 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 3157889 120 + 27905053 GCAGCUGAUCAAAGGCCCCCAAACUUGAUUGAAGUCUGAGGGACACUGACGAUUGCAGUGCAAAUGAUAAUUUAUUAGAUUGCGAUCUCUGGCCUGGCACAAAAAUGUGCCAGUCGCGGA ((.(((......(((((((((.((((.....)))).)).))......((.(((((((((.(.(((((....))))).)))))))))))).)))))((((((....))))))))).))... ( -40.80) >DroSec_CAF1 5743 120 + 1 GCAGCUGAUCAAAGGCCCCCAAACUUGAUUGAAGUCUGAGGGACACUGACGAUUGCAGUGCAAAUGAUAAUUUAUUAGAUUGCGAUCUCUGGCCUGGCACAAAAAUGUGUCAGUCGCGAA ((.(((......(((((((((.((((.....)))).)).))......((.(((((((((.(.(((((....))))).)))))))))))).)))))((((((....))))))))).))... ( -38.10) >DroSim_CAF1 7917 120 + 1 GCAGCUGAUCAAAGGCCCCCAAACUUGAUUGAAGUCUGAGGGACACUGACGAUUGCAGUGCAAAUGAUAAUUUAUUAGAUUGCGAUCUCUGGCCUGGCACAAAAAUGUGCCAGUCGCGAA ((.(((......(((((((((.((((.....)))).)).))......((.(((((((((.(.(((((....))))).)))))))))))).)))))((((((....))))))))).))... ( -40.80) >DroYak_CAF1 5447 116 + 1 GCAGCUGAUCAAAGGCCCCCAAACUUGAUUGAAGUCCGAGGGACGCUGACGAUUGCAGUUCAAAUGAUAAUUUAUUAGAUUGCGAUCUCUGGCAUGGCACAAAAAUGUGC----CGUGAA .((((.((((((.(.........)))))))...((((...))))))))..((((((((((..(((((....))))).)))))))))).....(((((((((....)))))----)))).. ( -39.10) >consensus GCAGCUGAUCAAAGGCCCCCAAACUUGAUUGAAGUCUGAGGGACACUGACGAUUGCAGUGCAAAUGAUAAUUUAUUAGAUUGCGAUCUCUGGCCUGGCACAAAAAUGUGCCAGUCGCGAA ((.(((..(((....(((.((.((((.....)))).)).)))....))).(((((((((.(.(((((....))))).))))))))))...)))((((((((....))))))))..))... (-35.67 = -36.30 + 0.63)

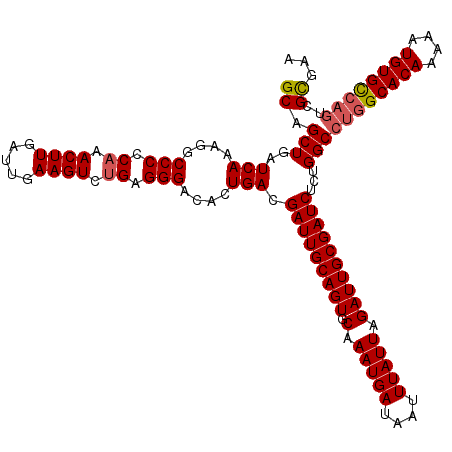
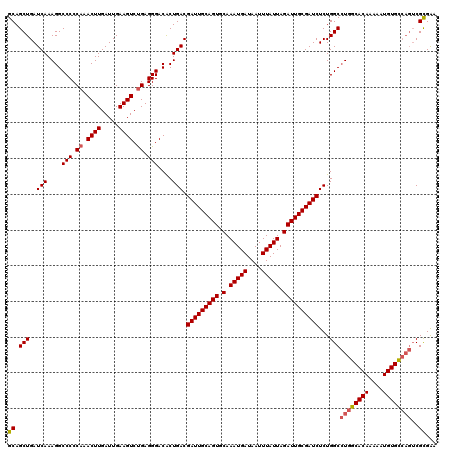
| Location | 3,157,889 – 3,158,009 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.42 |
| Mean single sequence MFE | -38.60 |
| Consensus MFE | -33.76 |
| Energy contribution | -35.07 |
| Covariance contribution | 1.31 |
| Combinations/Pair | 1.03 |
| Mean z-score | -3.05 |
| Structure conservation index | 0.87 |
| SVM decision value | 1.54 |
| SVM RNA-class probability | 0.962507 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 3157889 120 - 27905053 UCCGCGACUGGCACAUUUUUGUGCCAGGCCAGAGAUCGCAAUCUAAUAAAUUAUCAUUUGCACUGCAAUCGUCAGUGUCCCUCAGACUUCAAUCAAGUUUGGGGGCCUUUGAUCAGCUGC ...((..((((((((....))))))))......((((......................(((((((....).))))))(((((((((((.....))))))))))).....)))).))... ( -42.40) >DroSec_CAF1 5743 120 - 1 UUCGCGACUGACACAUUUUUGUGCCAGGCCAGAGAUCGCAAUCUAAUAAAUUAUCAUUUGCACUGCAAUCGUCAGUGUCCCUCAGACUUCAAUCAAGUUUGGGGGCCUUUGAUCAGCUGC ...((..(((.((((....)))).)))......((((......................(((((((....).))))))(((((((((((.....))))))))))).....)))).))... ( -35.30) >DroSim_CAF1 7917 120 - 1 UUCGCGACUGGCACAUUUUUGUGCCAGGCCAGAGAUCGCAAUCUAAUAAAUUAUCAUUUGCACUGCAAUCGUCAGUGUCCCUCAGACUUCAAUCAAGUUUGGGGGCCUUUGAUCAGCUGC ...((..((((((((....))))))))......((((......................(((((((....).))))))(((((((((((.....))))))))))).....)))).))... ( -42.40) >DroYak_CAF1 5447 116 - 1 UUCACG----GCACAUUUUUGUGCCAUGCCAGAGAUCGCAAUCUAAUAAAUUAUCAUUUGAACUGCAAUCGUCAGCGUCCCUCGGACUUCAAUCAAGUUUGGGGGCCUUUGAUCAGCUGC .....(----(((((....))))))....(((.(((.(((.((.(((........))).))..))).)))(((((.(.(((((((((((.....))))))))))).).)))))...))). ( -34.30) >consensus UUCGCGACUGGCACAUUUUUGUGCCAGGCCAGAGAUCGCAAUCUAAUAAAUUAUCAUUUGCACUGCAAUCGUCAGUGUCCCUCAGACUUCAAUCAAGUUUGGGGGCCUUUGAUCAGCUGC .......((((((((....))))))))..(((.((((......................(((((((....).))))))(((((((((((.....))))))))))).....))))..))). (-33.76 = -35.07 + 1.31)

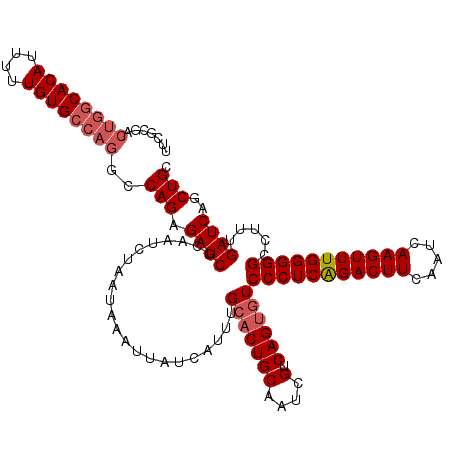
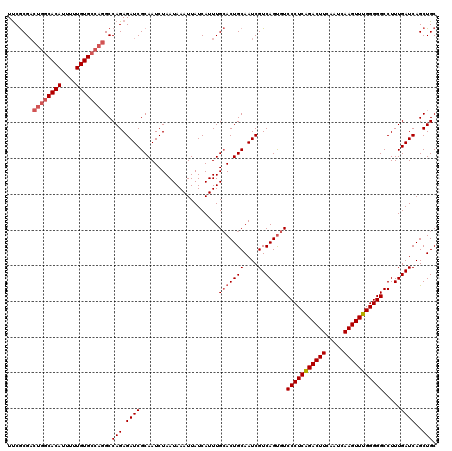
| Location | 3,157,929 – 3,158,049 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.00 |
| Mean single sequence MFE | -35.38 |
| Consensus MFE | -29.80 |
| Energy contribution | -30.42 |
| Covariance contribution | 0.63 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.18 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.73 |
| SVM RNA-class probability | 0.835935 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 3157929 120 + 27905053 GGACACUGACGAUUGCAGUGCAAAUGAUAAUUUAUUAGAUUGCGAUCUCUGGCCUGGCACAAAAAUGUGCCAGUCGCGGAUAGAAGAUCGGGGCUCCUUUGGCAUUUUUAUUGAAUUAAU (((..((((((((((((((.(.(((((....))))).))))))))))(((((.((((((((....)))))))).).)))).....).))))...)))....................... ( -36.30) >DroSec_CAF1 5783 120 + 1 GGACACUGACGAUUGCAGUGCAAAUGAUAAUUUAUUAGAUUGCGAUCUCUGGCCUGGCACAAAAAUGUGUCAGUCGCGAAUAGAAGAUCGGGGCUCCUUUGGCAUUUUUAUUGAAUUAAU .(.(((((.......))))))......(((((((.((((.(((((((((((((((((((((....))))))))..))...))).)))))((....))....)))..)))).))))))).. ( -32.50) >DroSim_CAF1 7957 120 + 1 GGACACUGACGAUUGCAGUGCAAAUGAUAAUUUAUUAGAUUGCGAUCUCUGGCCUGGCACAAAAAUGUGCCAGUCGCGAAUAGAAGAUCGGGGCUCCUUUGGCAUUUUUAUUGAAUUAAU .(.(((((.......))))))......(((((((.((((.(((((((((((((((((((((....))))))))..))...))).)))))((....))....)))..)))).))))))).. ( -35.20) >DroYak_CAF1 5487 116 + 1 GGACGCUGACGAUUGCAGUUCAAAUGAUAAUUUAUUAGAUUGCGAUCUCUGGCAUGGCACAAAAAUGUGC----CGUGAAUAGAAGCUCCGGGCUCCUUUGGCAUUUUUAUUGAAUUAAU (((.(((...((((((((((..(((((....))))).))))))))))((((.(((((((((....)))))----))))..))))))))))..(((.....)))................. ( -37.50) >consensus GGACACUGACGAUUGCAGUGCAAAUGAUAAUUUAUUAGAUUGCGAUCUCUGGCCUGGCACAAAAAUGUGCCAGUCGCGAAUAGAAGAUCGGGGCUCCUUUGGCAUUUUUAUUGAAUUAAU ((((.((((((((((((((.(.(((((....))))).))))))))))((((((((((((((....))))))))..))...)))).).)))).).)))....................... (-29.80 = -30.42 + 0.63)

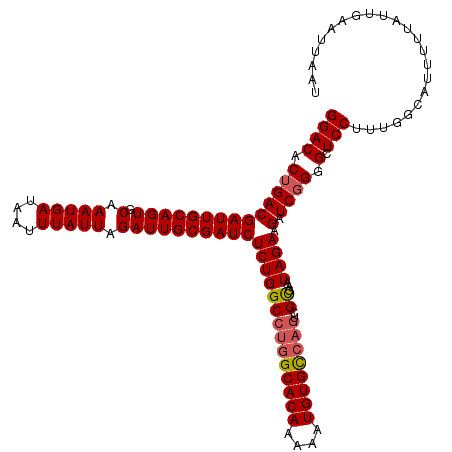
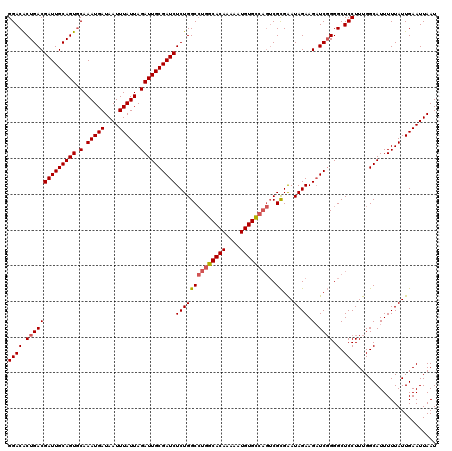
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:57:01 2006