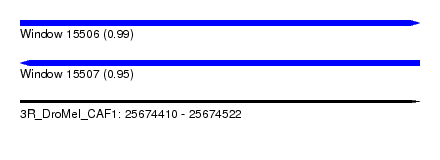
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 25,674,410 – 25,674,522 |
| Length | 112 |
| Max. P | 0.987506 |

| Location | 25,674,410 – 25,674,522 |
|---|---|
| Length | 112 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 78.51 |
| Mean single sequence MFE | -33.81 |
| Consensus MFE | -26.09 |
| Energy contribution | -26.78 |
| Covariance contribution | 0.70 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.22 |
| Structure conservation index | 0.77 |
| SVM decision value | 2.08 |
| SVM RNA-class probability | 0.987506 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 25674410 112 + 27905053 U--------GCAAGUGGCAGACUUGCUCCUUAACUUCGGCUGAGAAGCAGCCACCGUUAGUUACAUAACAGCCCAGCAGGCCUUUGUGAAGAGGAGAUGCACUGCAGCCACUUGACUCCG .--------.((((((((.......((((((......(((((.....)))))...........(((((..(((.....)))..)))))..)))))).(((...)))))))))))...... ( -39.60) >DroSim_CAF1 170 108 + 1 U--------GCAAGUGGCAAACUUGCUCCUUAACUUCGGCUGAGAAGCAGCCACCGUUAGUUACAUAACAGCCCAGCGGCCCUUUGUG---AGCAGAUGCACUGCAGCCACUUGACUCC- .--------.((((((((....(((((.((((.(....).)))).))))).(((.((((......)))).(((....))).....)))---.((((.....)))).)))))))).....- ( -36.00) >DroYak_CAF1 200 99 + 1 UCUCGUUCUUCAAGUGGCAAACUUGCUCCUUAACUUCGG-----------CCACCGU----UAUAUAACAGCCCAGCAGGCUUUUGUGA-----AGAUCCACUGCAGCCACUUGACUCC- .........(((((((((.....(((.......((((((-----------...))..----...((((.((((.....)))).))))))-----)).......))))))))))))....- ( -25.84) >consensus U________GCAAGUGGCAAACUUGCUCCUUAACUUCGGCUGAGAAGCAGCCACCGUUAGUUACAUAACAGCCCAGCAGGCCUUUGUGA__AG_AGAUGCACUGCAGCCACUUGACUCC_ ..........((((((((......(((..........(((((.....)))))...((((......)))))))...((((......................)))).))))))))...... (-26.09 = -26.78 + 0.70)

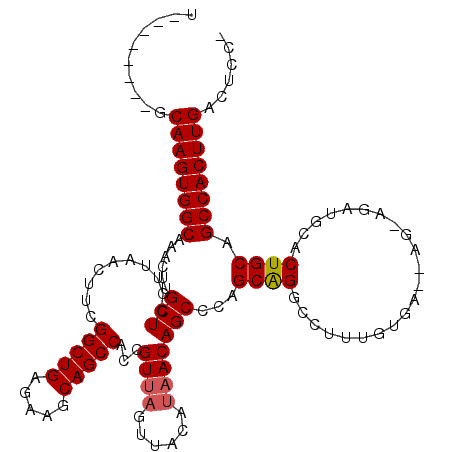
| Location | 25,674,410 – 25,674,522 |
|---|---|
| Length | 112 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 78.51 |
| Mean single sequence MFE | -37.83 |
| Consensus MFE | -28.42 |
| Energy contribution | -29.57 |
| Covariance contribution | 1.14 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.94 |
| Structure conservation index | 0.75 |
| SVM decision value | 1.40 |
| SVM RNA-class probability | 0.950538 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 25674410 112 - 27905053 CGGAGUCAAGUGGCUGCAGUGCAUCUCCUCUUCACAAAGGCCUGCUGGGCUGUUAUGUAACUAACGGUGGCUGCUUCUCAGCCGAAGUUAAGGAGCAAGUCUGCCACUUGC--------A ....(.((((((((.....(((...((((....(((..((((.....))))....)))...((((..((((((.....))))))..))))))))))).....)))))))))--------. ( -39.80) >DroSim_CAF1 170 108 - 1 -GGAGUCAAGUGGCUGCAGUGCAUCUGCU---CACAAAGGGCCGCUGGGCUGUUAUGUAACUAACGGUGGCUGCUUCUCAGCCGAAGUUAAGGAGCAAGUUUGCCACUUGC--------A -...(.((((((((.(((((.((.(.(((---(.....)))).).)).)))))..(((..(((((..((((((.....))))))..))).))..))).....)))))))))--------. ( -42.90) >DroYak_CAF1 200 99 - 1 -GGAGUCAAGUGGCUGCAGUGGAUCU-----UCACAAAAGCCUGCUGGGCUGUUAUAUA----ACGGUGG-----------CCGAAGUUAAGGAGCAAGUUUGCCACUUGAAGAACGAGA -....(((((((((....((((....-----))))..((((.((((..((((((....)----)))))..-----------((........)))))).)))))))))))))......... ( -30.80) >consensus _GGAGUCAAGUGGCUGCAGUGCAUCU_CU__UCACAAAGGCCUGCUGGGCUGUUAUGUAACUAACGGUGGCUGCUUCUCAGCCGAAGUUAAGGAGCAAGUUUGCCACUUGC________A ......((((((((.....(((.(((............(((((...)))))..........((((..((((((.....))))))..))))))).))).....)))))))).......... (-28.42 = -29.57 + 1.14)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:40:19 2006