
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 25,663,881 – 25,663,984 |
| Length | 103 |
| Max. P | 0.999655 |

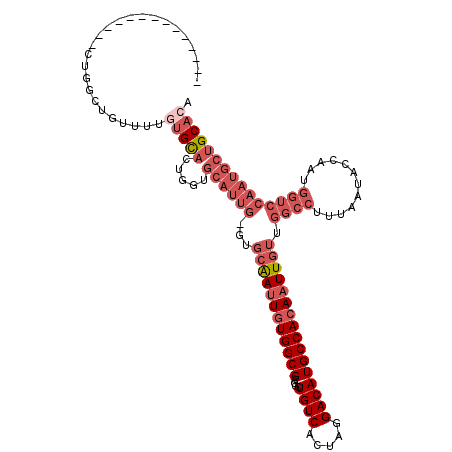
| Location | 25,663,881 – 25,663,984 |
|---|---|
| Length | 103 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 77.89 |
| Mean single sequence MFE | -34.79 |
| Consensus MFE | -22.88 |
| Energy contribution | -26.16 |
| Covariance contribution | 3.28 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.27 |
| Structure conservation index | 0.66 |
| SVM decision value | 1.79 |
| SVM RNA-class probability | 0.977346 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 25663881 103 + 27905053 --------------CUGGCUGUUUUGUGCACUGGUGCAUUG-GUGCAAUUGUGGCGAGGCUGUCACUAGGACAUGCCACAAUUGUUGGCCUUUAAUACCAAUGGUCCAAUGCUGCACA --------------..........((((((.....((((((-(.((((((((((((....((((.....))))))))))))))))..(((............)))))))))))))))) ( -36.90) >DroSec_CAF1 10361 103 + 1 --------------CUGGCUCUUUUGUGCACUGGUGCAUUG-GUGCAAUUGUGGCGAGGCUGUCACUAGGACAUGCCACAAUUGUUGGCCUUUAAUACCAAUGGUCCAAUGCUGCACA --------------..........((((((.....((((((-(.((((((((((((....((((.....))))))))))))))))..(((............)))))))))))))))) ( -36.90) >DroSim_CAF1 11291 103 + 1 --------------CUGGCUGUUUUGUGCACUGGUGCAUUG-GUGCAAUUGUGGCGAGGCUGUCACUAGGACAUGCCACAAUUGUUGGCCUUUAAUACCAAUGGUCCAAUGCUGCACA --------------..........((((((.....((((((-(.((((((((((((....((((.....))))))))))))))))..(((............)))))))))))))))) ( -36.90) >DroYak_CAF1 11153 114 + 1 GCAGCUAUUCCACAUUGGUACAUUGCUGCAUUGGUGCAUUG-GUGCAAUUGUGGCGAGGCUGUCACUAGGACAUGCCACAAUUGUUGGCCUUUAAUACCAAUGGUCCAAUGCUGC--- (((((.......((((((((......((((....))))..(-((((((((((((((....((((.....))))))))))))))))..))).....)))))))).......)))))--- ( -44.54) >DroAna_CAF1 10460 82 + 1 --------------------GUUUUGUGUUAU--UGUAUUUACUCGGAGUUUGGCGAGGCUGUCACUAGGACAUGCCACAAUUGUUGGCCUUUAAUGCC--------------GCAAA --------------------...(((((....--........((((........))))((((((.....)))).)))))))((((.(((.......)))--------------)))). ( -18.70) >consensus ______________CUGGCUGUUUUGUGCACUGGUGCAUUG_GUGCAAUUGUGGCGAGGCUGUCACUAGGACAUGCCACAAUUGUUGGCCUUUAAUACCAAUGGUCCAAUGCUGCACA .........................(((((.....((((((...((((((((((((....((((.....)))))))))))))))).((((............))))))))))))))). (-22.88 = -26.16 + 3.28)


| Location | 25,663,881 – 25,663,984 |
|---|---|
| Length | 103 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 77.89 |
| Mean single sequence MFE | -34.82 |
| Consensus MFE | -24.84 |
| Energy contribution | -26.86 |
| Covariance contribution | 2.02 |
| Combinations/Pair | 1.08 |
| Mean z-score | -3.66 |
| Structure conservation index | 0.71 |
| SVM decision value | 3.84 |
| SVM RNA-class probability | 0.999655 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 25663881 103 - 27905053 UGUGCAGCAUUGGACCAUUGGUAUUAAAGGCCAACAAUUGUGGCAUGUCCUAGUGACAGCCUCGCCACAAUUGCAC-CAAUGCACCAGUGCACAAAACAGCCAG-------------- (((((((((((((....(((((.......)))))((((((((((.((((.....)))).....))))))))))..)-)))))).....))))))..........-------------- ( -38.70) >DroSec_CAF1 10361 103 - 1 UGUGCAGCAUUGGACCAUUGGUAUUAAAGGCCAACAAUUGUGGCAUGUCCUAGUGACAGCCUCGCCACAAUUGCAC-CAAUGCACCAGUGCACAAAAGAGCCAG-------------- (((((((((((((....(((((.......)))))((((((((((.((((.....)))).....))))))))))..)-)))))).....))))))..........-------------- ( -38.70) >DroSim_CAF1 11291 103 - 1 UGUGCAGCAUUGGACCAUUGGUAUUAAAGGCCAACAAUUGUGGCAUGUCCUAGUGACAGCCUCGCCACAAUUGCAC-CAAUGCACCAGUGCACAAAACAGCCAG-------------- (((((((((((((....(((((.......)))))((((((((((.((((.....)))).....))))))))))..)-)))))).....))))))..........-------------- ( -38.70) >DroYak_CAF1 11153 114 - 1 ---GCAGCAUUGGACCAUUGGUAUUAAAGGCCAACAAUUGUGGCAUGUCCUAGUGACAGCCUCGCCACAAUUGCAC-CAAUGCACCAAUGCAGCAAUGUACCAAUGUGGAAUAGCUGC ---...(((((((....(((((.......)))))((((((((((.((((.....)))).....))))))))))..)-))))))......(((((..(((.((.....)).)))))))) ( -41.40) >DroAna_CAF1 10460 82 - 1 UUUGC--------------GGCAUUAAAGGCCAACAAUUGUGGCAUGUCCUAGUGACAGCCUCGCCAAACUCCGAGUAAAUACA--AUAACACAAAAC-------------------- .....--------------(((.......))).....(((((((.((((.....))))))((((........))))........--....)))))...-------------------- ( -16.60) >consensus UGUGCAGCAUUGGACCAUUGGUAUUAAAGGCCAACAAUUGUGGCAUGUCCUAGUGACAGCCUCGCCACAAUUGCAC_CAAUGCACCAGUGCACAAAACAGCCAG______________ ......(((((((....(((((.......)))))((((((((((.((((.....)))).....))))))))))...........)))))))........................... (-24.84 = -26.86 + 2.02)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:40:15 2006