| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 25,662,329 – 25,662,420 |
| Length | 91 |
| Max. P | 0.803915 |

| Location | 25,662,329 – 25,662,420 |
|---|---|
| Length | 91 |
| Sequences | 5 |
| Columns | 92 |
| Reading direction | forward |
| Mean pairwise identity | 82.11 |
| Mean single sequence MFE | -23.73 |
| Consensus MFE | -12.52 |
| Energy contribution | -14.52 |
| Covariance contribution | 2.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.42 |
| Structure conservation index | 0.53 |
| SVM decision value | 0.63 |
| SVM RNA-class probability | 0.803915 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 25662329 91 + 27905053 CUACACAAAAUUCAA-AGAAUGGGGCGAAAAAGGGGUCGAAAGGAGUAUUGUCCCCCCAUUUACAAGCGCCAGAAAGUGGCGCGCGUCACUA ...............-.((((((((.((........((....)).......)).))))))))....((((((.....))))))......... ( -27.26) >DroSec_CAF1 8775 88 + 1 CUGCACAAAAUUAAA-AGAAUGGGGCAAAAAAGACAUCGAAC---AUAUUGUCCCCCCACUUACAAGCGCCAGAAAGUGGCGCGCAUCACCA .(((...........-....(((((.......((((......---....)))).))))).......((((((.....)))))))))...... ( -22.50) >DroSim_CAF1 9706 88 + 1 CUGCACAAAAUUAAA-AGAAUGGGGCAAAAGAGACAUCGAAC---AUAUUGUCCCCCCACUUACAAGCGCCAGAAAGUGGCGCGCAUCACCA .(((...........-....(((((.....(.((((......---....)))))))))).......((((((.....)))))))))...... ( -23.30) >DroEre_CAF1 8839 91 + 1 UAACACAAAAUCCGA-GGAAUGGAGAGAAGAAAGCGUGGACAUAUGUAUUGUCCACCCAUUUACAAGAGCCAGAAAGUGGCGCGCAUCACCA ..........((...-.)).(((.((...(...(((((((((.......)))))))............((((.....)))))).).)).))) ( -21.40) >DroYak_CAF1 9633 92 + 1 UUACACAAAAUCCAAAAGAAUGGAGGGAAAAAAGCGUGGACAUGUGUAUUGUCCACCCAUUUACAAGAGCCAGAAAGUGGCGCGCAUCACCA ..........((((......))))((((.....(((((((((.......)))))))............((((.....))))))...)).)). ( -24.20) >consensus CUACACAAAAUUCAA_AGAAUGGGGCGAAAAAGGCGUCGAAA___GUAUUGUCCCCCCAUUUACAAGCGCCAGAAAGUGGCGCGCAUCACCA .................((((((((.((.......................)).))))))))....((((((.....))))))......... (-12.52 = -14.52 + 2.00)

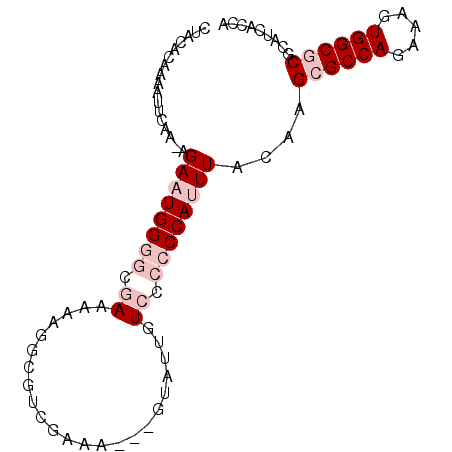
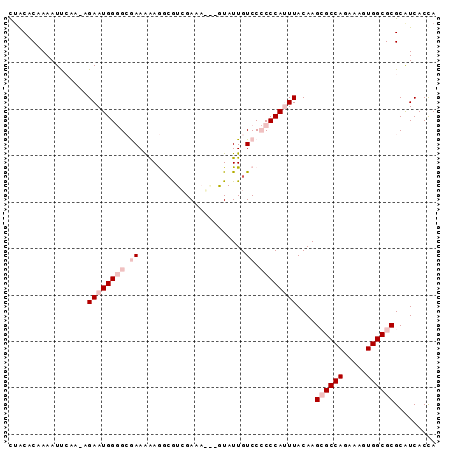
| Location | 25,662,329 – 25,662,420 |
|---|---|
| Length | 91 |
| Sequences | 5 |
| Columns | 92 |
| Reading direction | reverse |
| Mean pairwise identity | 82.11 |
| Mean single sequence MFE | -25.08 |
| Consensus MFE | -17.36 |
| Energy contribution | -16.80 |
| Covariance contribution | -0.56 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.56 |
| Structure conservation index | 0.69 |
| SVM decision value | -0.03 |
| SVM RNA-class probability | 0.518943 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 25662329 91 - 27905053 UAGUGACGCGCGCCACUUUCUGGCGCUUGUAAAUGGGGGGACAAUACUCCUUUCGACCCCUUUUUCGCCCCAUUCU-UUGAAUUUUGUGUAG .....((((((((((.....)))))).....((((((((((......)))...(((........))))))))))..-.........)))).. ( -26.90) >DroSec_CAF1 8775 88 - 1 UGGUGAUGCGCGCCACUUUCUGGCGCUUGUAAGUGGGGGGACAAUAU---GUUCGAUGUCUUUUUUGCCCCAUUCU-UUUAAUUUUGUGCAG ......(((((((((.....)))))).....(((((((((((.....---))))(....).......)))))))..-...........))). ( -25.60) >DroSim_CAF1 9706 88 - 1 UGGUGAUGCGCGCCACUUUCUGGCGCUUGUAAGUGGGGGGACAAUAU---GUUCGAUGUCUCUUUUGCCCCAUUCU-UUUAAUUUUGUGCAG ......(((((((((.....)))))).....(((((((((((.....---))))((....)).....)))))))..-...........))). ( -26.20) >DroEre_CAF1 8839 91 - 1 UGGUGAUGCGCGCCACUUUCUGGCUCUUGUAAAUGGGUGGACAAUACAUAUGUCCACGCUUUCUUCUCUCCAUUCC-UCGGAUUUUGUGUUA ...((((((((((((.....))))............(((((((.......))))))))).........(((.....-..)))....)))))) ( -21.80) >DroYak_CAF1 9633 92 - 1 UGGUGAUGCGCGCCACUUUCUGGCUCUUGUAAAUGGGUGGACAAUACACAUGUCCACGCUUUUUUCCCUCCAUUCUUUUGGAUUUUGUGUAA ......(((((((((.....))))..........(.(((((((.......))))))).).........((((......))))....))))). ( -24.90) >consensus UGGUGAUGCGCGCCACUUUCUGGCGCUUGUAAAUGGGGGGACAAUAC___UUUCGACGCCUUUUUCGCCCCAUUCU_UUGAAUUUUGUGUAG .......((((((((.....)))).......(((((((.((.......................)).)))))))............)))).. (-17.36 = -16.80 + -0.56)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:40:13 2006