
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 25,644,215 – 25,644,364 |
| Length | 149 |
| Max. P | 0.838137 |

| Location | 25,644,215 – 25,644,326 |
|---|---|
| Length | 111 |
| Sequences | 3 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 82.81 |
| Mean single sequence MFE | -35.02 |
| Consensus MFE | -28.81 |
| Energy contribution | -29.03 |
| Covariance contribution | 0.23 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.74 |
| SVM RNA-class probability | 0.838137 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
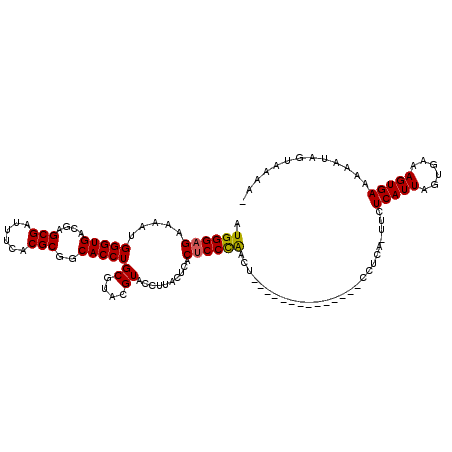
>3R_DroMel_CAF1 25644215 111 + 27905053 GGUGGAUUUGGCCUAAGCCGAUUAGGCUAGAAUGGGAGAAAAUGGGUGACGAGCGAUUUCACGCGACACCUGCGUACGUACCUUUCUCACUCCCAACUCCCAACUCCACAC .(((((((((((((((.....)))))))))).((((((.....((((((.(((.......(((((.....)))))........)))))))))....))))))..))))).. ( -41.36) >DroSim_CAF1 11152 98 + 1 GGUGGAUUUGGCCUAAGCCGAUUAGGCUACAAUGGGAGAAAAUGGGUGACGAGCGAUUUCACGCGGCACCUGCGUACGUACCUUCCUCACUCCCAACU------------- ........((((((((.....))))))))...((((((.....(((((.((...(....)((((((...)))))).)))))))......))))))...------------- ( -32.00) >DroYak_CAF1 11127 95 + 1 GGUGGAUUUGGCCUAAGCCGAUUAGGCUAGAAUGGGAGAAAAUGGGUGACGAGCGAUUUCACGCGGCACCUGCGUACGUAUUGUACAUACACCUG---------------- ((((..((((((((((.....))))))))))............(((((....(((......)))..)))))..((((.....))))...))))..---------------- ( -31.70) >consensus GGUGGAUUUGGCCUAAGCCGAUUAGGCUAGAAUGGGAGAAAAUGGGUGACGAGCGAUUUCACGCGGCACCUGCGUACGUACCUUACUCACUCCCAACU_____________ ......((((((((((.....)))))))))).((((((.....(((((....(((......)))..)))))((....))..........))))))................ (-28.81 = -29.03 + 0.23)


| Location | 25,644,246 – 25,644,364 |
|---|---|
| Length | 118 |
| Sequences | 3 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 78.36 |
| Mean single sequence MFE | -25.39 |
| Consensus MFE | -19.67 |
| Energy contribution | -19.57 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.27 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.10 |
| SVM RNA-class probability | 0.584641 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 25644246 118 + 27905053 AUGGGAGAAAAUGGGUGACGAGCGAUUUCACGCGACACCUGCGUACGUACCUUUCUCACUCCCAACUCCCAACUCCACACUCCUCAUUUCUCAUUAGUGAAAGUGAAAAAUAGUAAAA- .((((((.....((((((.(((.......(((((.....)))))........)))))))))....))))))......((((..(((((.......))))).)))).............- ( -28.16) >DroSim_CAF1 11183 104 + 1 AUGGGAGAAAAUGGGUGACGAGCGAUUUCACGCGGCACCUGCGUACGUACCUUCCUCACUCCCAACU--------------CCUCACUUCUCAUUAGUGAAAGUGAAAAAUAGUAAAA- .((((((.....(((((.((...(....)((((((...)))))).)))))))......))))))...--------------..((((((.(((....)))))))))............- ( -28.90) >DroYak_CAF1 11158 95 + 1 AUGGGAGAAAAUGGGUGACGAGCGAUUUCACGCGGCACCUGCGUACGUAUUGUACAUACACCUG------------------------GCUCAUUAGUGAAAGUGAAAAAUAGUAAAAA ....(((....((((((..(.(((((...((((((...))))))....))))).)...))))))------------------------.)))........................... ( -19.10) >consensus AUGGGAGAAAAUGGGUGACGAGCGAUUUCACGCGGCACCUGCGUACGUACCUUACUCACUCCCAACU______________CCUCA_UUCUCAUUAGUGAAAGUGAAAAAUAGUAAAA_ .((((((.....(((((....(((......)))..)))))((....))..........))))))..........................(((((......)))))............. (-19.67 = -19.57 + -0.11)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:40:02 2006