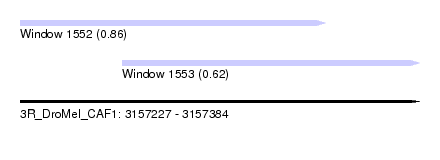
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 3,157,227 – 3,157,384 |
| Length | 157 |
| Max. P | 0.864723 |

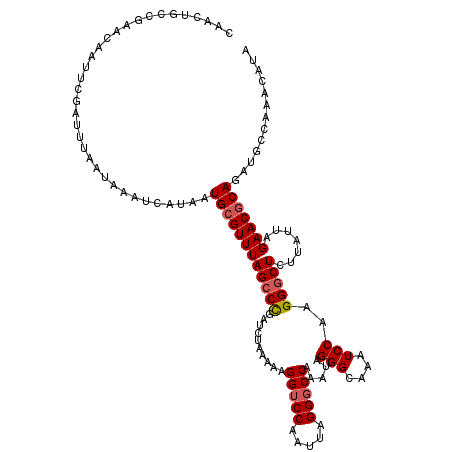
| Location | 3,157,227 – 3,157,347 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.67 |
| Mean single sequence MFE | -25.55 |
| Consensus MFE | -20.97 |
| Energy contribution | -21.54 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.84 |
| SVM RNA-class probability | 0.864723 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 3157227 120 + 27905053 CAACUGCCGAACAAUUCGAUUUAAUAAAUCAUAAUGCGUUUAGCCUGAUCUAAAAAGGUCCAAUUAGGGCCAAAUAGGGCAAAUCCAAGGGCUCUUAUUAGAACGCAGAUGCCAAACAUA .....((..........((((.....))))....((((((((((((..........(((((.....))))).....(((....)))..))))).......)))))))...))........ ( -25.21) >DroSec_CAF1 5082 120 + 1 CAACUGCCGAACAAUUCGAUUUAAUAAAUCAUAAUGCGUUUAGCCCGAUCUAAAAAGGUCCAAUUAGGGCCAAAUAGGGCAAAUCCAAGGGCUCUUAUUAGAACGCAGAUGCCAAACAUA .....((..........((((.....))))....((((((((((((..........(((((.....))))).....(((....)))..))))).......)))))))...))........ ( -27.81) >DroSim_CAF1 7226 120 + 1 CAACUGCCGAACAAUUCGAUUUAAUAAAUCAUAAUGCGUUUAGCCCGAUCUAAAAAGGUCCAAUUAGGCCCAAAUAGGGCAAAUCCAAGGGCUCUUAUUAGAACGCAGAUGCCAAACAUA .....((..........((((.....))))....(((((((((((((((((....))))).......((((.....))))........))))).......)))))))...))........ ( -29.21) >DroYak_CAF1 4777 104 + 1 CAAC----------------UUAAUAAAUCAUAAUGCGUUUAGCCCGAUCUAAAAAGGUCCAAUUAGGGCCAAAUAGGGCAAAUCCAAGGUCUCUUAUUAGAACUCAGAUGCCAAACAUA ....----------------.................((((.((((..........(((((.....))))).....))))........(((.(((...........))).)))))))... ( -19.96) >consensus CAACUGCCGAACAAUUCGAUUUAAUAAAUCAUAAUGCGUUUAGCCCGAUCUAAAAAGGUCCAAUUAGGGCCAAAUAGGGCAAAUCCAAGGGCUCUUAUUAGAACGCAGAUGCCAAACAUA ..................................((((((((((((..........(((((.....))))).....(((....)))..))))).......)))))))............. (-20.97 = -21.54 + 0.56)

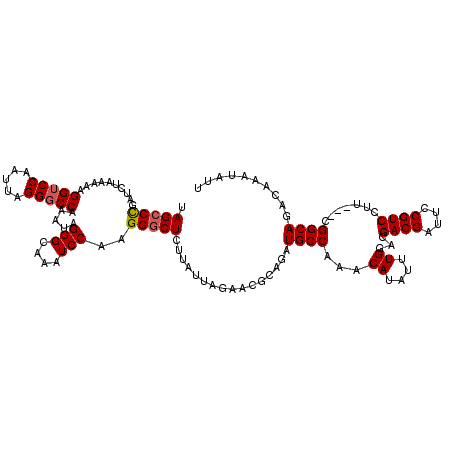
| Location | 3,157,267 – 3,157,384 |
|---|---|
| Length | 117 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.80 |
| Mean single sequence MFE | -31.53 |
| Consensus MFE | -25.06 |
| Energy contribution | -25.38 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.06 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.17 |
| SVM RNA-class probability | 0.617776 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 3157267 117 + 27905053 UAGCCUGAUCUAAAAAGGUCCAAUUAGGGCCAAAUAGGGCAAAUCCAAGGGCUCUUAUUAGAACGCAGAUGCCAAACAUAUUUGGACGACGAUGCCGUCCUU---CGGCAGACAAAUAUU ....(((.(((((.(((((((.......(((......)))........)))).))).)))))...))).(((((((....))))).))....(((((.....---))))).......... ( -29.46) >DroSec_CAF1 5122 117 + 1 UAGCCCGAUCUAAAAAGGUCCAAUUAGGGCCAAAUAGGGCAAAUCCAAGGGCUCUUAUUAGAACGCAGAUGCCAAACAUAUUUGGACGACGAUUCCGUCCUU---CGGCAGACAAAUAUU .(((((..........(((((.....))))).....(((....)))..)))))................(((((((....)))(((((.......)))))..---.)))).......... ( -29.50) >DroSim_CAF1 7266 117 + 1 UAGCCCGAUCUAAAAAGGUCCAAUUAGGCCCAAAUAGGGCAAAUCCAAGGGCUCUUAUUAGAACGCAGAUGCCAAACAUAUUUGGCCGACGAUUCCGUCCUU---CGGCAGACAAAUAUU ..(((((.(((((.(((((((......((((.....))))........)))).))).))))).))..((.((((((....)))))).((((....))))..)---))))........... ( -34.94) >DroYak_CAF1 4801 120 + 1 UAGCCCGAUCUAAAAAGGUCCAAUUAGGGCCAAAUAGGGCAAAUCCAAGGUCUCUUAUUAGAACUCAGAUGCCAAACAUAUUUGCCCGACGAUUCCGUCCUCCGCCGGCACACAAAUAUU ..(((.(((((....))))).......(((......((((((((....(((.(((...........))).)))......))))))))((((....))))....))))))........... ( -32.20) >consensus UAGCCCGAUCUAAAAAGGUCCAAUUAGGGCCAAAUAGGGCAAAUCCAAGGGCUCUUAUUAGAACGCAGAUGCCAAACAUAUUUGGACGACGAUUCCGUCCUU___CGGCAGACAAAUAUU .(((((..........(((((.....))))).....(((....)))..)))))................((((...((....))...((((....)))).......)))).......... (-25.06 = -25.38 + 0.31)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:56:55 2006