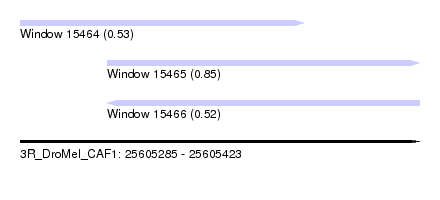
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 25,605,285 – 25,605,423 |
| Length | 138 |
| Max. P | 0.850374 |

| Location | 25,605,285 – 25,605,383 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 77.50 |
| Mean single sequence MFE | -29.83 |
| Consensus MFE | -18.45 |
| Energy contribution | -19.40 |
| Covariance contribution | 0.95 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.33 |
| Structure conservation index | 0.62 |
| SVM decision value | -0.00 |
| SVM RNA-class probability | 0.531066 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 25605285 98 + 27905053 CCCGGCUGGCGCCCACAAGUAGAUCGUGCU----------CUCCACGCUCCUUUAGGCCAUUCAAUAUACUGAGCAACAUGCUCUCGGGCAUGUUAUCGAACAAGCCA ...((((.(((((...(((..((.((((..----------...)))).)))))..)))...(((......)))))((((((((....))))))))........)))). ( -27.80) >DroVir_CAF1 43003 101 + 1 AACGAUUGGCGGCCUGCUGCAGAUACUCG-GCACGAGUCUCGCCCUGCA------GCGCUCGCAACUGAUUGAUCAGCAUGCUCUCGGGCACAUUGUCGAAGAGGCCG .........(((((((((((((..(((((-...)))))......)))))------))..((((((((((....))))..((((....))))..))).)))..)))))) ( -35.70) >DroSec_CAF1 32282 98 + 1 CCCGGCUGGCGCCCUCCAGGAGAUCGCGCU----------CUCCACGCUCCUUUAGGCCAUUCAAUAUACUGAGCAACAUGCUCUCGGGCAUGUUGUCGAACAAGCCA ...(((((((........(((((......)----------))))...((.....)))))...........(((.(((((((((....))))))))))))....)))). ( -31.90) >DroSim_CAF1 33091 98 + 1 CCCGGCUGGCGCCCUCCAGGAGAUCGCGCU----------CUCCACGCUCCUUUAGGCCAUUCAAUAUACUGAGCAACGUGCUCUCGGGCAUGUUGUCGAACAAGCCA ...(((((((........(((((......)----------))))...((.....)))))...........(((.(((((((((....))))))))))))....)))). ( -31.50) >DroEre_CAF1 33250 98 + 1 CCUGGCUGGCGCACUGCAGCAGAUCCCGCU----------CCCCACGCUCCUUUAAGCCAUUCAAUAUCUUGAGCAACAUGCUCUCGGGCAUGUUAUCGAACAAUCCA ..(((((((.((...(.(((.......)))----------.)....)).))....))))).........((((..((((((((....)))))))).))))........ ( -24.40) >DroYak_CAF1 34384 98 + 1 CCUGGCUGGCGCCCUGCAGCAGAUCCCGCU----------CCCCACGCUCCUUCAAACCAUUCAAUAUCUUGAGCAACAUGCUCUCGGGCAUGUUGUCGAACAAGCCA ..(((..((((..(((...)))....))))----------..))).(((..(((.......((((....))))((((((((((....)))))))))).)))..))).. ( -27.70) >consensus CCCGGCUGGCGCCCUCCAGCAGAUCCCGCU__________CUCCACGCUCCUUUAGGCCAUUCAAUAUACUGAGCAACAUGCUCUCGGGCAUGUUGUCGAACAAGCCA ...(((((((((.(((...)))...)))))........................................(((.(((((((((....))))))))))))....)))). (-18.45 = -19.40 + 0.95)
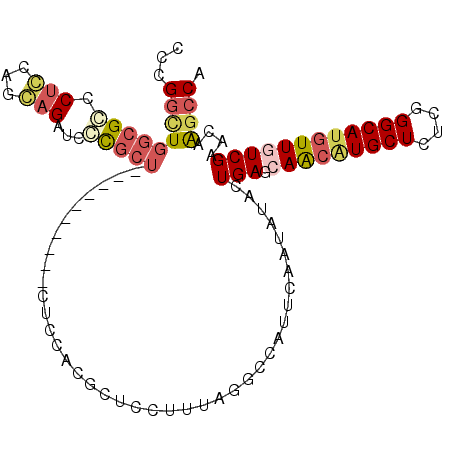

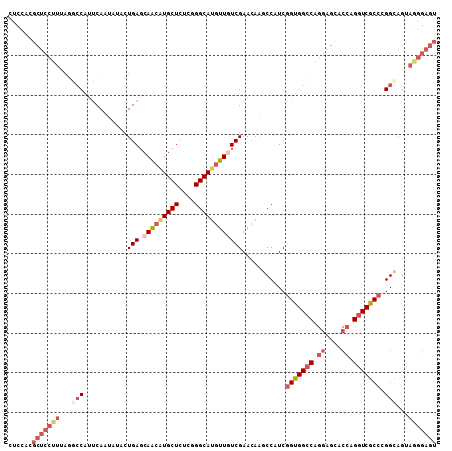
| Location | 25,605,315 – 25,605,423 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 83.27 |
| Mean single sequence MFE | -43.77 |
| Consensus MFE | -31.17 |
| Energy contribution | -33.53 |
| Covariance contribution | 2.36 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.93 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.79 |
| SVM RNA-class probability | 0.850374 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 25605315 108 + 27905053 CUCCACGCUCCUUUAGGCCAUUCAAUAUACUGAGCAACAUGCUCUCGGGCAUGUUAUCGAACAAGCCAUCGGUGGCCAGGAGCACCAGGUCACCGGGUAGUAGGGAGU ((((..(((((....(((..((((......)))).((((((((....)))))))).........)))..((((((((.((....)).)))))))))).)))..)))). ( -42.90) >DroVir_CAF1 43042 102 + 1 CGCCCUGCA------GCGCUCGCAACUGAUUGAUCAGCAUGCUCUCGGGCACAUUGUCGAAGAGGCCGUCCGUGGCCAGCAGUACAAGAUCGCCCGGCUGUAGUGGCA .((((((((------((....(((.((((....))))..)))...(((((...(((((.....(((((....)))))....).))))....)))))))))))).))). ( -43.60) >DroSec_CAF1 32312 108 + 1 CUCCACGCUCCUUUAGGCCAUUCAAUAUACUGAGCAACAUGCUCUCGGGCAUGUUGUCGAACAAGCCAUCGGUGGCCAGGAGCACCAGGUCGCCGGGUAGUAGGGAGU ((((..(((((....(((...(((......)))((((((((((....)))))))))).......)))..((((((((.((....)).)))))))))).)))..)))). ( -46.10) >DroSim_CAF1 33121 108 + 1 CUCCACGCUCCUUUAGGCCAUUCAAUAUACUGAGCAACGUGCUCUCGGGCAUGUUGUCGAACAAGCCAUCGGUGGCCAGGAGCACCAGGUCGCCGGGUAGUAGGGAGU ((((..(((((....(((...(((......)))((((((((((....)))))))))).......)))..((((((((.((....)).)))))))))).)))..)))). ( -45.70) >DroEre_CAF1 33280 108 + 1 CCCCACGCUCCUUUAAGCCAUUCAAUAUCUUGAGCAACAUGCUCUCGGGCAUGUUAUCGAACAAUCCAUCGGUGGCCAGGAGCACCAGGUCGCCCGGCAGCAAGGAGU ......(((((((...(((..........((((..((((((((....)))))))).))))..........(((((((.((....)).))))))).)))...))))))) ( -41.20) >DroYak_CAF1 34414 108 + 1 CCCCACGCUCCUUCAAACCAUUCAAUAUCUUGAGCAACAUGCUCUCGGGCAUGUUGUCGAACAAGCCAUCGGUGGCCAGGAGCACCAGGUCGCCCGGCAGCAAGGAGU ......((((((((...............((((.(((((((((....)))))))))))))....(((...(((((((.((....)).))))))).))).).))))))) ( -43.10) >consensus CUCCACGCUCCUUUAGGCCAUUCAAUAUACUGAGCAACAUGCUCUCGGGCAUGUUGUCGAACAAGCCAUCGGUGGCCAGGAGCACCAGGUCGCCCGGCAGUAGGGAGU ......(((((((...(((...........(((.(((((((((....))))))))))))...........(((((((.((....)).))))))).)))...))))))) (-31.17 = -33.53 + 2.36)

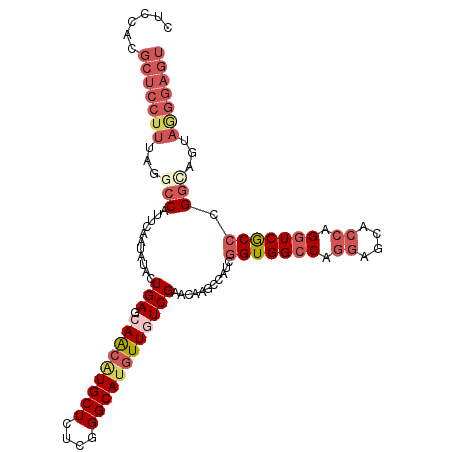
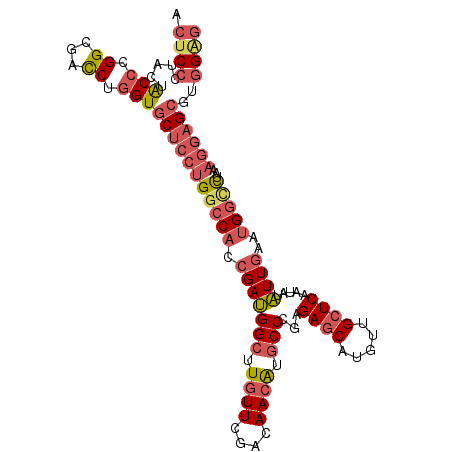
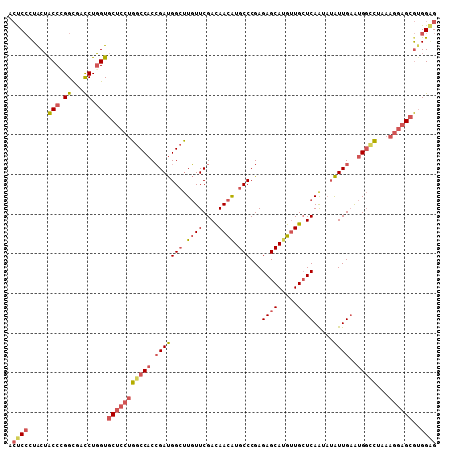
| Location | 25,605,315 – 25,605,423 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 83.27 |
| Mean single sequence MFE | -42.37 |
| Consensus MFE | -27.43 |
| Energy contribution | -28.80 |
| Covariance contribution | 1.37 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.95 |
| Structure conservation index | 0.65 |
| SVM decision value | -0.03 |
| SVM RNA-class probability | 0.518771 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 25605315 108 - 27905053 ACUCCCUACUACCCGGUGACCUGGUGCUCCUGGCCACCGAUGGCUUGUUCGAUAACAUGCCCGAGAGCAUGUUGCUCAGUAUAUUGAAUGGCCUAAAGGAGCGUGGAG .(((((....(((.(....)..)))(((((((((((.(((((((.((((....)))).)))...((((.....)))).....))))..)))))...))))))).)))) ( -40.10) >DroVir_CAF1 43042 102 - 1 UGCCACUACAGCCGGGCGAUCUUGUACUGCUGGCCACGGACGGCCUCUUCGACAAUGUGCCCGAGAGCAUGCUGAUCAAUCAGUUGCGAGCGC------UGCAGGGCG .(((.((.((((((((((...((((......((((......))))......))))..))))))...(((.(((((....))))))))....))------)).))))). ( -43.30) >DroSec_CAF1 32312 108 - 1 ACUCCCUACUACCCGGCGACCUGGUGCUCCUGGCCACCGAUGGCUUGUUCGACAACAUGCCCGAGAGCAUGUUGCUCAGUAUAUUGAAUGGCCUAAAGGAGCGUGGAG .(((((....(((.((...)).)))(((((((((((.(((((..(((...(.((((((((......))))))))).)))..)))))..)))))...))))))).)))) ( -40.50) >DroSim_CAF1 33121 108 - 1 ACUCCCUACUACCCGGCGACCUGGUGCUCCUGGCCACCGAUGGCUUGUUCGACAACAUGCCCGAGAGCACGUUGCUCAGUAUAUUGAAUGGCCUAAAGGAGCGUGGAG .(((((....(((.((...)).)))(((((((((((.(((((((.((((....)))).)))...((((.....)))).....))))..)))))...))))))).)))) ( -39.80) >DroEre_CAF1 33280 108 - 1 ACUCCUUGCUGCCGGGCGACCUGGUGCUCCUGGCCACCGAUGGAUUGUUCGAUAACAUGCCCGAGAGCAUGUUGCUCAAGAUAUUGAAUGGCUUAAAGGAGCGUGGGG ...(((..(.((((((...))))))(((((((((((.(((((..(((...(.((((((((......))))))))).)))..)))))..)))))...)))))))..))) ( -44.80) >DroYak_CAF1 34414 108 - 1 ACUCCUUGCUGCCGGGCGACCUGGUGCUCCUGGCCACCGAUGGCUUGUUCGACAACAUGCCCGAGAGCAUGUUGCUCAAGAUAUUGAAUGGUUUGAAGGAGCGUGGGG ...(((..(.((((((...))))))((((((.((((.(((((.((((...(.((((((((......))))))))).)))).)))))..))))....)))))))..))) ( -45.70) >consensus ACUCCCUACUACCCGGCGACCUGGUGCUCCUGGCCACCGAUGGCUUGUUCGACAACAUGCCCGAGAGCAUGUUGCUCAAUAUAUUGAAUGGCCUAAAGGAGCGUGGAG .((((.....(((.((...)).)))(((((((((((.(((((((.((((....)))).)))...((((.....)))).....))))..)))))...))))))..)))) (-27.43 = -28.80 + 1.37)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:39:42 2006