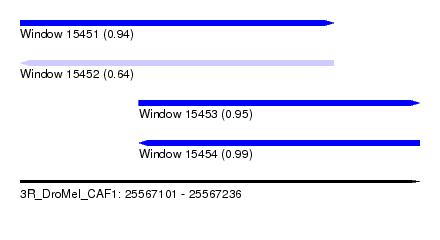
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 25,567,101 – 25,567,236 |
| Length | 135 |
| Max. P | 0.987035 |

| Location | 25,567,101 – 25,567,207 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 80.85 |
| Mean single sequence MFE | -29.15 |
| Consensus MFE | -20.51 |
| Energy contribution | -20.90 |
| Covariance contribution | 0.39 |
| Combinations/Pair | 1.20 |
| Mean z-score | -2.69 |
| Structure conservation index | 0.70 |
| SVM decision value | 1.24 |
| SVM RNA-class probability | 0.935214 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 25567101 106 + 27905053 AUUUUGCCUCUUGACAAUUAGCCGCCAACGCUGCUCCAAAGUCCAAGCUUUGAGUGGGAGCACC--CCAUCUCAAAGUU-A---CAAAUGCGCGAAGCGCAAUGCAAC-AA--AA ...((((.....(((....(((.((....)).))).....)))..(((((((((((((.....)--))).)))))))))-.---....(((((...)))))..)))).-..--.. ( -31.30) >DroVir_CAF1 59206 101 + 1 AUUUUGCCUCUUGACAAUUAGCCGCCAACGCUGCUCCAAAAUCCAAGCUUUAG-AUCAUCCAAU--CUAUCU-AAAGCA-------ACUGCGCGAGGCGCAAUGUAAAA-C--CA (((.((((((.((.((...(((.((....)).)))...........(((((((-((........--..))))-))))).-------..)))).)))))).)))......-.--.. ( -24.10) >DroPse_CAF1 49045 106 + 1 AUUUUGCCUCUUGACAAUUAGCCGCCAACGCUGCUCCAAAAUCCAAGCUUUGAGUUGAUC-CCU--CAACCUCAAGGCU-G---CAACUGCGCGAGGCGCAAUGUAAAGCA--AA .....(((((.((.((...(((.((....)).)))..........(((((((((((((..-..)--))).)))))))))-.---....)))).)))))((........)).--.. ( -30.30) >DroGri_CAF1 60967 107 + 1 AAUUUGCCUCUUGACAAUUAGCCGCCAACGCUGCUCCAAAAUCCAAGCUUUAGGAUAAUC--CU--CAACCU-AAAGCU-ACACAAACUGCGCGAGGCGCAAUGUAAACAA--CA .....(((((.((.((...(((.((....)).)))..........(((((((((......--..--...)))-))))))-........)))).))))).............--.. ( -25.40) >DroAna_CAF1 60261 111 + 1 AUUUUGCCUCUUGACAAUUAGCCGCCAACGCUGCUCCAAAGUCCAAGCUUUGAGUGAGAUUACCACUCAUCUCAAGGCUGA---AAAGUGCGCGAGGCGCAAUGCAGA-AUAUCA (((((((.....(((....(((.((....)).))).....)))..(((((((((((((.......)))).)))))))))..---....(((((...)))))..)))))-)).... ( -33.50) >DroPer_CAF1 48090 106 + 1 AUUUUGCCUCUUGACAAUUAGCCGCCAACGCUGCUCCAAAAUCCAAGCUUUGAGUUGAUC-CCU--CAACCUCAAGGCU-G---CAACUGCGCGAGGCGCAAUGUAAAGCA--AA .....(((((.((.((...(((.((....)).)))..........(((((((((((((..-..)--))).)))))))))-.---....)))).)))))((........)).--.. ( -30.30) >consensus AUUUUGCCUCUUGACAAUUAGCCGCCAACGCUGCUCCAAAAUCCAAGCUUUGAGUUGAUC_ACU__CAACCUCAAAGCU_A___CAACUGCGCGAGGCGCAAUGUAAA_AA__AA .....(((((.((.((...(((.((....)).)))...........(((((((((((.........))).))))))))..........)))).)))))................. (-20.51 = -20.90 + 0.39)



| Location | 25,567,101 – 25,567,207 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 80.85 |
| Mean single sequence MFE | -34.03 |
| Consensus MFE | -21.76 |
| Energy contribution | -22.46 |
| Covariance contribution | 0.69 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.22 |
| SVM RNA-class probability | 0.638216 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 25567101 106 - 27905053 UU--UU-GUUGCAUUGCGCUUCGCGCAUUUG---U-AACUUUGAGAUGG--GGUGCUCCCACUCAAAGCUUGGACUUUGGAGCAGCGUUGGCGGCUAAUUGUCAAGAGGCAAAAU ..--..-((((((.(((((...)))))..))---)-)))((((((.(((--(.....))))))))))((((.(((.....(((.((....)).)))....)))...))))..... ( -37.30) >DroVir_CAF1 59206 101 - 1 UG--G-UUUUACAUUGCGCCUCGCGCAGU-------UGCUUU-AGAUAG--AUUGGAUGAU-CUAAAGCUUGGAUUUUGGAGCAGCGUUGGCGGCUAAUUGUCAAGAGGCAAAAU ..--.-...........(((((..((.((-------((((((-((((((--(((....)))-)))..........)))))))))))....))(((.....)))..)))))..... ( -30.00) >DroPse_CAF1 49045 106 - 1 UU--UGCUUUACAUUGCGCCUCGCGCAGUUG---C-AGCCUUGAGGUUG--AGG-GAUCAACUCAAAGCUUGGAUUUUGGAGCAGCGUUGGCGGCUAAUUGUCAAGAGGCAAAAU ..--.((........))(((((..(((((((---(-.((((((((.(((--(..-..))))))))).((((.(....).))))......))).)).))))))...)))))..... ( -35.90) >DroGri_CAF1 60967 107 - 1 UG--UUGUUUACAUUGCGCCUCGCGCAGUUUGUGU-AGCUUU-AGGUUG--AG--GAUUAUCCUAAAGCUUGGAUUUUGGAGCAGCGUUGGCGGCUAAUUGUCAAGAGGCAAAUU .(--((((..((.((((.((..((((.....))))-((((((-(((.((--(.--..))).)))))))))........)).)))).))..)))))...(((((....)))))... ( -32.00) >DroAna_CAF1 60261 111 - 1 UGAUAU-UCUGCAUUGCGCCUCGCGCACUUU---UCAGCCUUGAGAUGAGUGGUAAUCUCACUCAAAGCUUGGACUUUGGAGCAGCGUUGGCGGCUAAUUGUCAAGAGGCAAAAU ......-..........(((((((((.((.(---(((((.(((((.((((.......))))))))).)).))))....)).)).)).(((((((....))))))))))))..... ( -33.10) >DroPer_CAF1 48090 106 - 1 UU--UGCUUUACAUUGCGCCUCGCGCAGUUG---C-AGCCUUGAGGUUG--AGG-GAUCAACUCAAAGCUUGGAUUUUGGAGCAGCGUUGGCGGCUAAUUGUCAAGAGGCAAAAU ..--.((........))(((((..(((((((---(-.((((((((.(((--(..-..))))))))).((((.(....).))))......))).)).))))))...)))))..... ( -35.90) >consensus UG__UU_UUUACAUUGCGCCUCGCGCAGUUG___U_AGCCUUGAGAUUG__AGU_GAUCAACUCAAAGCUUGGAUUUUGGAGCAGCGUUGGCGGCUAAUUGUCAAGAGGCAAAAU .................(((((((((..............(((((................))))).((((.(....).)))).))))((((((....)))))).)))))..... (-21.76 = -22.46 + 0.69)



| Location | 25,567,141 – 25,567,236 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 74.69 |
| Mean single sequence MFE | -26.05 |
| Consensus MFE | -24.97 |
| Energy contribution | -23.72 |
| Covariance contribution | -1.25 |
| Combinations/Pair | 1.27 |
| Mean z-score | -3.66 |
| Structure conservation index | 0.96 |
| SVM decision value | 1.35 |
| SVM RNA-class probability | 0.945014 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 25567141 95 + 27905053 GUCCAAGCUUUGAGUGGGAGCACCCCAUCUCAAAGUUA---CAAAUGCGCGAAGCGCAAUGCAAC-AAAACU-AAAAUUGGAUUCUAAGAAAACAUUAAA ((((((((((((((((((.....)))).))))))))).---....(((((...))))).......-......-.....)))))................. ( -28.00) >DroPse_CAF1 49085 95 + 1 AUCCAAGCUUUGAGUUGAUC-CCUCAACCUCAAGGCUG---CAACUGCGCGAGGCGCAAUGUAAAGCAAACG-AAAAUUGGACCCAAACAAAAGAAAACU .(((((((((((((((((..-..)))).))))))))((---((..(((((...))))).)))).........-....))))).................. ( -26.40) >DroGri_CAF1 61007 92 + 1 AUCCAAGCUUUAGGAUAAUC--CUCAACCU-AAAGCUACACAAACUGCGCGAGGCGCAAUGUAAACAACACAAUCAAAUGGGCUCGA-----ACAUAAAA .(((((((((((((......--.....)))-))))))..(((...(((((...))))).)))................)))).....-----........ ( -20.60) >DroSec_CAF1 49377 95 + 1 GUCCAAGCUUUGAGUGGGUCCACCCCAUCUCAAAGUUA---CAAAUGCGCGAAGCGCAAUGCAAC-AACACU-GAAAUUGGAUUCUAAGAAAACAUAAAA ((((((((((((((((((.....)))).))))))))).---....(((((...))))).......-......-.....)))))................. ( -27.50) >DroEre_CAF1 50214 95 + 1 GUCCAAGCUUUGAGUGGGUGCACCCCAGCUCAAAGUUA---UAAGUGCGCGAAGCGCAAUGCAAG-AAAACU-GAAAUUGGAUUCUAACAAAACAUAAAA ((((((((((((((((((.....)))).))))))))).---....(((((...))))).......-......-.....)))))................. ( -27.40) >DroPer_CAF1 48130 95 + 1 AUCCAAGCUUUGAGUUGAUC-CCUCAACCUCAAGGCUG---CAACUGCGCGAGGCGCAAUGUAAAGCAAACG-AAAAUUGGACCCAAACAAAAGAAAACU .(((((((((((((((((..-..)))).))))))))((---((..(((((...))))).)))).........-....))))).................. ( -26.40) >consensus AUCCAAGCUUUGAGUGGAUC_ACCCAACCUCAAAGCUA___CAACUGCGCGAAGCGCAAUGCAAA_AAAACU_AAAAUUGGACUCUAACAAAACAUAAAA .(((((((((((((((((.....)))).)))))))))........(((((...)))))....................)))).................. (-24.97 = -23.72 + -1.25)


| Location | 25,567,141 – 25,567,236 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 74.69 |
| Mean single sequence MFE | -26.62 |
| Consensus MFE | -19.42 |
| Energy contribution | -19.45 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.11 |
| Structure conservation index | 0.73 |
| SVM decision value | 2.06 |
| SVM RNA-class probability | 0.987035 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 25567141 95 - 27905053 UUUAAUGUUUUCUUAGAAUCCAAUUUU-AGUUUU-GUUGCAUUGCGCUUCGCGCAUUUG---UAACUUUGAGAUGGGGUGCUCCCACUCAAAGCUUGGAC ..................(((((....-......-((((((.(((((...)))))..))---))))((((((.((((.....))))))))))..))))). ( -28.90) >DroPse_CAF1 49085 95 - 1 AGUUUUCUUUUGUUUGGGUCCAAUUUU-CGUUUGCUUUACAUUGCGCCUCGCGCAGUUG---CAGCCUUGAGGUUGAGG-GAUCAACUCAAAGCUUGGAU .................((((((....-....(((.....(((((((...))))))).)---))((.(((((.((((..-..))))))))).)))))))) ( -25.70) >DroGri_CAF1 61007 92 - 1 UUUUAUGU-----UCGAGCCCAUUUGAUUGUGUUGUUUACAUUGCGCCUCGCGCAGUUUGUGUAGCUUU-AGGUUGAG--GAUUAUCCUAAAGCUUGGAU ......((-----((((((............((((..((.(((((((...))))))).))..))))(((-(((.(((.--..))).)))))))))))))) ( -23.60) >DroSec_CAF1 49377 95 - 1 UUUUAUGUUUUCUUAGAAUCCAAUUUC-AGUGUU-GUUGCAUUGCGCUUCGCGCAUUUG---UAACUUUGAGAUGGGGUGGACCCACUCAAAGCUUGGAC ..................(((((....-......-((((((.(((((...)))))..))---))))((((((.((((.....))))))))))..))))). ( -30.10) >DroEre_CAF1 50214 95 - 1 UUUUAUGUUUUGUUAGAAUCCAAUUUC-AGUUUU-CUUGCAUUGCGCUUCGCGCACUUA---UAACUUUGAGCUGGGGUGCACCCACUCAAAGCUUGGAC ..(((((...(((.((((..(......-.)..))-)).))).(((((...)))))..))---)))(((((((.((((.....)))))))))))....... ( -25.70) >DroPer_CAF1 48130 95 - 1 AGUUUUCUUUUGUUUGGGUCCAAUUUU-CGUUUGCUUUACAUUGCGCCUCGCGCAGUUG---CAGCCUUGAGGUUGAGG-GAUCAACUCAAAGCUUGGAU .................((((((....-....(((.....(((((((...))))))).)---))((.(((((.((((..-..))))))))).)))))))) ( -25.70) >consensus UUUUAUGUUUUGUUAGAAUCCAAUUUU_AGUUUU_UUUACAUUGCGCCUCGCGCAGUUG___UAACUUUGAGGUGGAGU_GACCAACUCAAAGCUUGGAC ..................(((((...................(((((...)))))..........(((((((.((((.....))))))))))).))))). (-19.42 = -19.45 + 0.03)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:39:32 2006