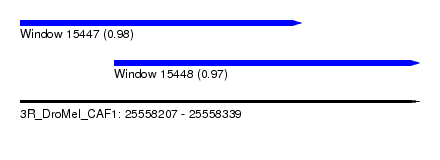
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 25,558,207 – 25,558,339 |
| Length | 132 |
| Max. P | 0.975495 |

| Location | 25,558,207 – 25,558,300 |
|---|---|
| Length | 93 |
| Sequences | 4 |
| Columns | 93 |
| Reading direction | forward |
| Mean pairwise identity | 92.83 |
| Mean single sequence MFE | -35.70 |
| Consensus MFE | -33.00 |
| Energy contribution | -33.62 |
| Covariance contribution | 0.62 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.19 |
| Structure conservation index | 0.92 |
| SVM decision value | 1.75 |
| SVM RNA-class probability | 0.975495 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
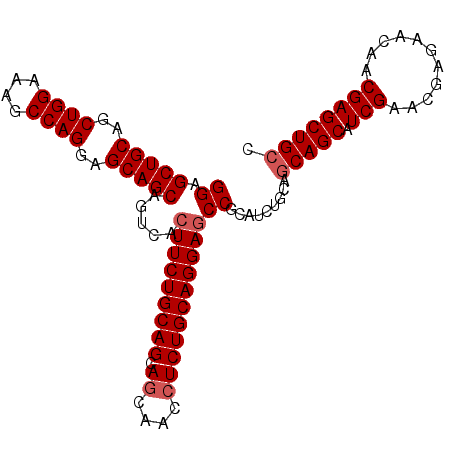
>3R_DroMel_CAF1 25558207 93 + 27905053 CCACAACUUCAGGACGCGCUCCACAGGCACAGGGGCUGCAGCUGGAAAGCCAGGAGCAGCAGUCACUUCUGCAGCAGCAACCUCUGCAGGAGC ............((((((((((.(((.(.....).)))..(((....)))..))))).)).))).(((((((((.((....))))))))))). ( -35.60) >DroSec_CAF1 41858 93 + 1 CCACAACUUCAGGACGCGCUCCACAGGCACAGGAGCUGCAGCUGGAAAGCCAGGAGCAGCAGUCCCUUCUGCAGCAGCAACCUCUGCAGGAGC ...........(((((((((((.........)))))(((..((((....))))..))))).))))(((((((((.((....))))))))))). ( -37.30) >DroSim_CAF1 42838 93 + 1 CCACAACUUCAGGACGCGCUCCACAGGCACAGGAGCUGCAGCUGGAAAGCCAGGAGCAGCAGUCCCUUCUGCAGCAGCAACCUCUGCAGGAGC ...........(((((((((((.........)))))(((..((((....))))..))))).))))(((((((((.((....))))))))))). ( -37.30) >DroYak_CAF1 45696 93 + 1 GAACAACUUCAGGUCGCGCUCUACGGGCACAGGAGCUGCAACUGGAAAGCCAGGAGCAGCAGGCACUUCUGCAGCAGUUACAUCUGCAGGAUC ...........(((((((((.....)))......(((((..((((....))))..)))))..))....((((((.(......))))))))))) ( -32.60) >consensus CCACAACUUCAGGACGCGCUCCACAGGCACAGGAGCUGCAGCUGGAAAGCCAGGAGCAGCAGUCACUUCUGCAGCAGCAACCUCUGCAGGAGC ...........(((((.(((.....))).)....(((((..((((....))))..))))).))))(((((((((.((....))))))))))). (-33.00 = -33.62 + 0.62)


| Location | 25,558,238 – 25,558,339 |
|---|---|
| Length | 101 |
| Sequences | 4 |
| Columns | 101 |
| Reading direction | forward |
| Mean pairwise identity | 94.88 |
| Mean single sequence MFE | -39.70 |
| Consensus MFE | -36.33 |
| Energy contribution | -37.07 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.44 |
| Structure conservation index | 0.91 |
| SVM decision value | 1.72 |
| SVM RNA-class probability | 0.973838 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 25558238 101 + 27905053 GGGGCUGCAGCUGGAAAGCCAGGAGCAGCAGUCACUUCUGCAGCAGCAACCUCUGCAGGAGCCUCAUCUGCAGCAGCAUCGAACGAGAACAACGAGCUGCC ((((((((..((((....))))..))))).....(((((((((.((....))))))))))).))).......(((((.(((...........)))))))). ( -40.20) >DroSec_CAF1 41889 101 + 1 GGAGCUGCAGCUGGAAAGCCAGGAGCAGCAGUCCCUUCUGCAGCAGCAACCUCUGCAGGAGCCGCAUCUGCAGCAGCAUCGAACGAGAACAACGAGCUGCC ((.(((((..((((....))))..))))).....(((((((((.((....)))))))))))))((....)).(((((.(((...........)))))))). ( -41.20) >DroSim_CAF1 42869 101 + 1 GGAGCUGCAGCUGGAAAGCCAGGAGCAGCAGUCCCUUCUGCAGCAGCAACCUCUGCAGGAGCCGCAUCUGCAGCAGCAUCGAACGAGAACAACGAGCUGCC ((.(((((..((((....))))..))))).....(((((((((.((....)))))))))))))((....)).(((((.(((...........)))))))). ( -41.20) >DroYak_CAF1 45727 101 + 1 GGAGCUGCAACUGGAAAGCCAGGAGCAGCAGGCACUUCUGCAGCAGUUACAUCUGCAGGAUCCGCAUCUGCAUCAGCAUCGAACGAGAACAACGAGCUGCC ((((((((..((((....))))..)))))......((((((((.(......))))))))))))((....))..((((.(((...........))))))).. ( -36.20) >consensus GGAGCUGCAGCUGGAAAGCCAGGAGCAGCAGUCACUUCUGCAGCAGCAACCUCUGCAGGAGCCGCAUCUGCAGCAGCAUCGAACGAGAACAACGAGCUGCC ((.(((((..((((....))))..))))).....(((((((((.((....))))))))))))).........(((((.(((...........)))))))). (-36.33 = -37.07 + 0.75)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:39:27 2006