
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
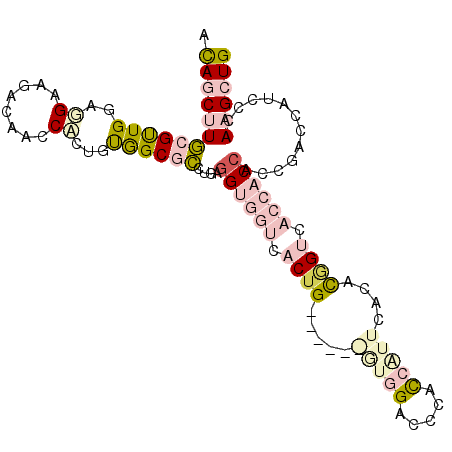
| Location | 25,548,436 – 25,548,540 |
| Length | 104 |
| Max. P | 0.934087 |

| Location | 25,548,436 – 25,548,540 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 75.28 |
| Mean single sequence MFE | -34.98 |
| Consensus MFE | -14.28 |
| Energy contribution | -16.65 |
| Covariance contribution | 2.37 |
| Combinations/Pair | 1.41 |
| Mean z-score | -2.09 |
| Structure conservation index | 0.41 |
| SVM decision value | 0.31 |
| SVM RNA-class probability | 0.681045 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 25548436 104 + 27905053 AUAGCUUGCGUUGGAGGAAGACAACCACUGUGGCGCCUGAAGUGGUCACUGGAUCCAGUGGAUCAACCGUUAACACGGUCACCACCACCGACCAUCCCAAGCUG .(((((((.(((((.((..((..((((((..(....)...))))))(((((....)))))..)).(((((....))))).....)).))))).....))))))) ( -35.10) >DroPse_CAF1 29658 95 + 1 ACAGCCUUCGCUGGCGGAAAACAACCCCAGCAGCCACAGAGGCAACCACUG---------GACACCCCAUGGACCUGGCCACCUCCACGGAGCAUCCCAAGAUG ...(((...(((((.((.......))))))).(((.....)))..(((.((---------(.....))))))....)))...(((....)))((((....)))) ( -28.60) >DroSec_CAF1 32036 104 + 1 ACAGCUUGCGUUGGAGGAAGACAACCAGUGUGGCGCCUGAGGUGGUCACUGGAUCCAGUGGACCCACCAUUCACACUGUCACCACCACCGACCAUCCCAAGCUG .(((((((.(((((.((........((((((((.......(((((((((((....))))))..)))))..))))))))......)).))))).....))))))) ( -40.04) >DroEre_CAF1 32872 101 + 1 ACAGCUUGCGUUGGAAGAAGACAACCUCUGUGGCGCCUGAGGUGGUCACUGCU---GAUGGACCCACCGUUCACACAGUCACCACCACCGACCAUCCCAAGCUG .(((((((.((((........))))....((((((...(.((((((.((((.(---(.(((((.....))))))))))).)))))).))).))))..))))))) ( -36.50) >DroYak_CAF1 35174 104 + 1 ACAGCUUGCGUUGGAAGAAGACAACCUCUGUGGCGCCGGAGGUGGUCACUGCUUCCGGUGGGGCCAUCAUUCACACAGUCACCACCACCGACCAUCCCAAGCUG .(((((((....(((((..(((.(((((((......))))))).)))....)))))((((((((.............))).)))))...........))))))) ( -41.02) >DroPer_CAF1 29228 95 + 1 ACAGCCUUCGCUGGCGGAAAACAACCCCAGCAGCCACAGAGGCAACCACUG---------GACACCCCAUGGACCUGGCCACCUCCACGGAGCAUCCCAAGAUG ...(((...(((((.((.......))))))).(((.....)))..(((.((---------(.....))))))....)))...(((....)))((((....)))) ( -28.60) >consensus ACAGCUUGCGUUGGAGGAAGACAACCACUGUGGCGCCUGAGGUGGUCACUG______GUGGACCCACCAUUCACACGGUCACCACCACCGACCAUCCCAAGCUG .((((((((((((..((........))...))))))....((((((.((((.....(((((.....)))))....)))).))))))............)))))) (-14.28 = -16.65 + 2.37)


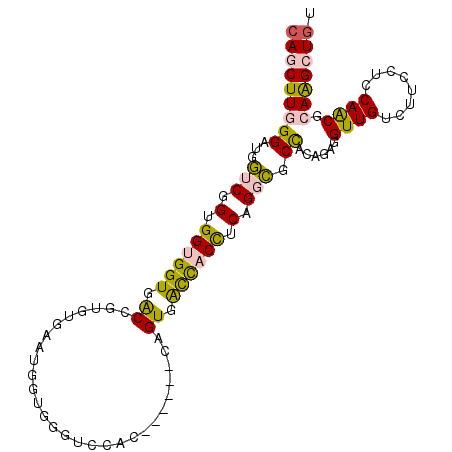
| Location | 25,548,436 – 25,548,540 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 75.28 |
| Mean single sequence MFE | -43.82 |
| Consensus MFE | -20.04 |
| Energy contribution | -19.41 |
| Covariance contribution | -0.63 |
| Combinations/Pair | 1.36 |
| Mean z-score | -2.31 |
| Structure conservation index | 0.46 |
| SVM decision value | 1.23 |
| SVM RNA-class probability | 0.934087 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 25548436 104 - 27905053 CAGCUUGGGAUGGUCGGUGGUGGUGACCGUGUUAACGGUUGAUCCACUGGAUCCAGUGACCACUUCAGGCGCCACAGUGGUUGUCUUCCUCCAACGCAAGCUAU .((((((.(.(((..((.((((((((((((....))))))....(((((....)))))))))))..(((((((.....)).))))).)).))).).)))))).. ( -42.90) >DroPse_CAF1 29658 95 - 1 CAUCUUGGGAUGCUCCGUGGAGGUGGCCAGGUCCAUGGGGUGUC---------CAGUGGUUGCCUCUGUGGCUGCUGGGGUUGUUUUCCGCCAGCGAAGGCUGU .......((((((((((((((..(....)..)))))))))))))---------).((((..(((((.((....)).)))))......))))((((....)))). ( -40.90) >DroSec_CAF1 32036 104 - 1 CAGCUUGGGAUGGUCGGUGGUGGUGACAGUGUGAAUGGUGGGUCCACUGGAUCCAGUGACCACCUCAGGCGCCACACUGGUUGUCUUCCUCCAACGCAAGCUGU (((((((.(.(((...((((((....).((.(((..(((((...(((((....))))).)))))))).)))))))...((.......)).))).).))))))). ( -45.80) >DroEre_CAF1 32872 101 - 1 CAGCUUGGGAUGGUCGGUGGUGGUGACUGUGUGAACGGUGGGUCCAUC---AGCAGUGACCACCUCAGGCGCCACAGAGGUUGUCUUCUUCCAACGCAAGCUGU (((((((.(.(((..((.((((((.((((((....)((((....))))---.))))).)))))).)(((((((.....)).))))).)..))).).))))))). ( -45.10) >DroYak_CAF1 35174 104 - 1 CAGCUUGGGAUGGUCGGUGGUGGUGACUGUGUGAAUGAUGGCCCCACCGGAAGCAGUGACCACCUCCGGCGCCACAGAGGUUGUCUUCUUCCAACGCAAGCUGU (((((((.(.(((((((.((((((.(((((.....((.((....)).))...))))).)))))).))))).....(((((....)))))..)).).))))))). ( -47.30) >DroPer_CAF1 29228 95 - 1 CAUCUUGGGAUGCUCCGUGGAGGUGGCCAGGUCCAUGGGGUGUC---------CAGUGGUUGCCUCUGUGGCUGCUGGGGUUGUUUUCCGCCAGCGAAGGCUGU .......((((((((((((((..(....)..)))))))))))))---------).((((..(((((.((....)).)))))......))))((((....)))). ( -40.90) >consensus CAGCUUGGGAUGGUCGGUGGUGGUGACCGUGUGAAUGGUGGGUCCAC______CAGUGACCACCUCAGGCGCCACAGAGGUUGUCUUCCUCCAACGCAAGCUGU (((((((((...(((.(.((((((.((............................)).)))))).).))).))......((((........)))).))))))). (-20.04 = -19.41 + -0.63)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:39:21 2006