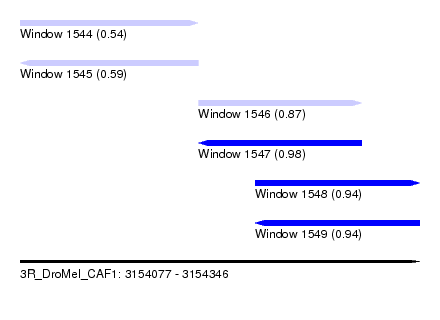
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 3,154,077 – 3,154,346 |
| Length | 269 |
| Max. P | 0.984307 |

| Location | 3,154,077 – 3,154,197 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.19 |
| Mean single sequence MFE | -34.97 |
| Consensus MFE | -31.48 |
| Energy contribution | -31.16 |
| Covariance contribution | -0.31 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.37 |
| Structure conservation index | 0.90 |
| SVM decision value | 0.02 |
| SVM RNA-class probability | 0.542141 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 3154077 120 + 27905053 GGGGCCAACGGCAAAUGGCAACAAAUUAGCCAAAGUGAUGAUGCGUUAAGUGGCAUAGUGGGGAAAUCGCCAGUGAGGAAAUUAGUUUUGGCGGUUUGCGAACCUAACGCACAGGGUGUU (((..(..(.((...((....)).....((((...(((((...)))))..))))...)).)(.((((((((((..............)))))))))).))..)))(((((.....))))) ( -32.94) >DroSec_CAF1 1965 120 + 1 GGGGCCAACGGCAAAUGGCAACAAAUUAGCCAAAGUGAUGAUGCGUUAAGUGGCAAAGUGGGGAAAUCGCCAGUGAGGAAAUUAGUUUUGGCGGUUGGCGGCGCAAGCACACAGGGUGUU ...((((((.((...((....))(((((((((...(((((...)))))..))))..(.(((.(....).))).)........)))))...)).)))))).((....))((((...)))). ( -36.00) >DroSim_CAF1 4135 120 + 1 GGGGCCAACGGCAAAUGGCAACAAAUUAGCCAAAGUGAUGAUGCGUUAAGUGGCAAAGUGGGGAAAUCGCCAGUAAGGAAAUUAGUUUUGGCGGUUGGCGGCGCAAGCACACAGGGUGUU ...((((((.((...((....))(((((((((...(((((...)))))..))))..(.(((.(....).))).)........)))))...)).)))))).((....))((((...)))). ( -36.00) >DroYak_CAF1 1265 118 + 1 GGGGCCAACGGCAAAUGGCAACAAAUUAGCCAAAGUGAUGAUGCGUUAAGUGGCAAAGUGGUGCAAUCGCCAGUGAGGAAAUUAGUUUUGGCGGUUGGCAGGG--AGCACACUUGGUGUU ...((((((.(((..((....)).....((((...(((((...)))))..)))).......)))...((((((..............))))))))))))....--(((((.....))))) ( -34.94) >consensus GGGGCCAACGGCAAAUGGCAACAAAUUAGCCAAAGUGAUGAUGCGUUAAGUGGCAAAGUGGGGAAAUCGCCAGUGAGGAAAUUAGUUUUGGCGGUUGGCGGCGCAAGCACACAGGGUGUU ...((((((......((....)).....(((((((((((....(.(((..((((..............)))).))).)..)))).))))))).))))))......(((((.....))))) (-31.48 = -31.16 + -0.31)

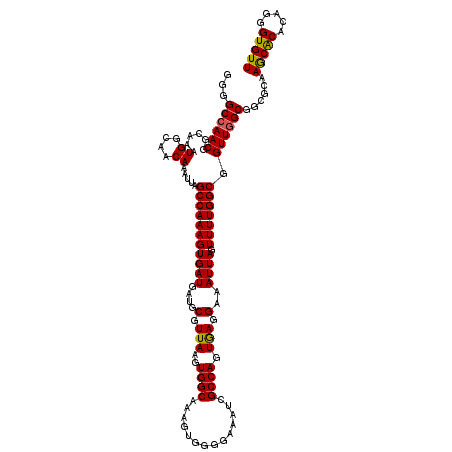
| Location | 3,154,077 – 3,154,197 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.19 |
| Mean single sequence MFE | -25.64 |
| Consensus MFE | -20.83 |
| Energy contribution | -21.57 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.10 |
| SVM RNA-class probability | 0.585688 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 3154077 120 - 27905053 AACACCCUGUGCGUUAGGUUCGCAAACCGCCAAAACUAAUUUCCUCACUGGCGAUUUCCCCACUAUGCCACUUAACGCAUCAUCACUUUGGCUAAUUUGUUGCCAUUUGCCGUUGGCCCC ........(((((((((((..(((((.(((((................))))).)).........))).))))))))))).........(((((((..((........)).))))))).. ( -27.49) >DroSec_CAF1 1965 120 - 1 AACACCCUGUGUGCUUGCGCCGCCAACCGCCAAAACUAAUUUCCUCACUGGCGAUUUCCCCACUUUGCCACUUAACGCAUCAUCACUUUGGCUAAUUUGUUGCCAUUUGCCGUUGGCCCC ........((((....)))).((((((.((..................((((((..........))))))..................((((.........))))...)).))))))... ( -25.40) >DroSim_CAF1 4135 120 - 1 AACACCCUGUGUGCUUGCGCCGCCAACCGCCAAAACUAAUUUCCUUACUGGCGAUUUCCCCACUUUGCCACUUAACGCAUCAUCACUUUGGCUAAUUUGUUGCCAUUUGCCGUUGGCCCC ........((((....)))).((((((.((..................((((((..........))))))..................((((.........))))...)).))))))... ( -25.40) >DroYak_CAF1 1265 118 - 1 AACACCAAGUGUGCU--CCCUGCCAACCGCCAAAACUAAUUUCCUCACUGGCGAUUGCACCACUUUGCCACUUAACGCAUCAUCACUUUGGCUAAUUUGUUGCCAUUUGCCGUUGGCCCC .((((...))))...--....(((((((((((................)))))...(((......(((........))).........((((.........))))..))).))))))... ( -24.29) >consensus AACACCCUGUGUGCUUGCGCCGCCAACCGCCAAAACUAAUUUCCUCACUGGCGAUUUCCCCACUUUGCCACUUAACGCAUCAUCACUUUGGCUAAUUUGUUGCCAUUUGCCGUUGGCCCC .((((...)))).........((((((.((..................((((((..........))))))..................((((.........))))...)).))))))... (-20.83 = -21.57 + 0.75)

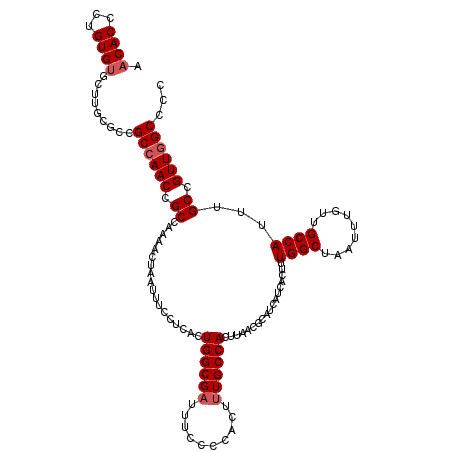
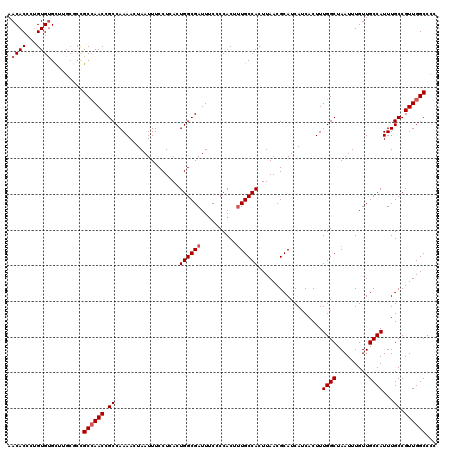
| Location | 3,154,197 – 3,154,307 |
|---|---|
| Length | 110 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 87.61 |
| Mean single sequence MFE | -26.12 |
| Consensus MFE | -23.02 |
| Energy contribution | -23.90 |
| Covariance contribution | 0.88 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.78 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.86 |
| SVM RNA-class probability | 0.867985 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 3154197 110 + 27905053 AGGUCUUUCAAGGGAACGA--CACUCGUCACAUUGCUGUGACAGCUGCCCACACUCCUGGCCACUUUGUCUUUGGCCAAUUAAAAUCAAAUUUCAAAAUU--------UCAAAAUGUUCC ............((((((.--.....((((((....))))))...............((((((.........))))))......................--------......)))))) ( -22.30) >DroSec_CAF1 2085 104 + 1 AGGUCUUUCAAGGGAACGGCGCACUCGUCACAUUGCUGUGACAGCUGCCCACACUCCUGGCCACUUUGUCUUUGGCCAAUUAAAAUCAAAUUU----------------CAAAAUGUUCC ............(((((((.(((((.((((((....)))))))).))))).......((((((.........))))))...............----------------......))))) ( -29.10) >DroSim_CAF1 4255 104 + 1 AGGUCUUUCAAGGGAACGGCGCACUCGUCACAUUGCUGUGACAGCUGCCCACACUCCUGGCCACUUUGUCUUUGGCCAAUUAAAAUCAAAUUU----------------CAAAAUGUUCC ............(((((((.(((((.((((((....)))))))).))))).......((((((.........))))))...............----------------......))))) ( -29.10) >DroYak_CAF1 1383 118 + 1 AGGUCUUUCAGAGGAACGG--CACUCGUCACAUUGCUGUGACAGCACCCCACACUCCUGGGCACUUUGUCUUUGCCCAAUUAAAAUCAAAUUUCAAAAUUUCAGCAUUUCAAAAUGUUCC ............(((((((--..((.((((((....))))))))..)).........((((((.........)))))).....................................))))) ( -24.00) >consensus AGGUCUUUCAAGGGAACGG__CACUCGUCACAUUGCUGUGACAGCUGCCCACACUCCUGGCCACUUUGUCUUUGGCCAAUUAAAAUCAAAUUU________________CAAAAUGUUCC ............(((((((.(((((.((((((....)))))))).))))).......((((((.........)))))).....................................))))) (-23.02 = -23.90 + 0.88)

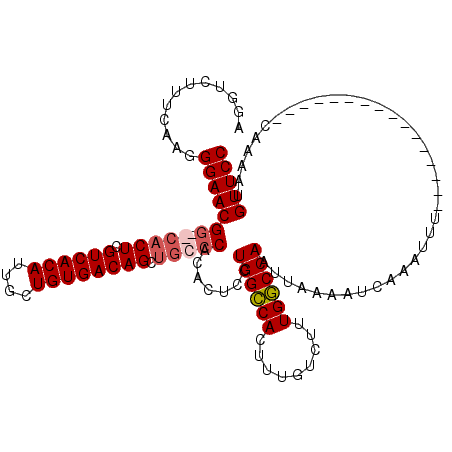
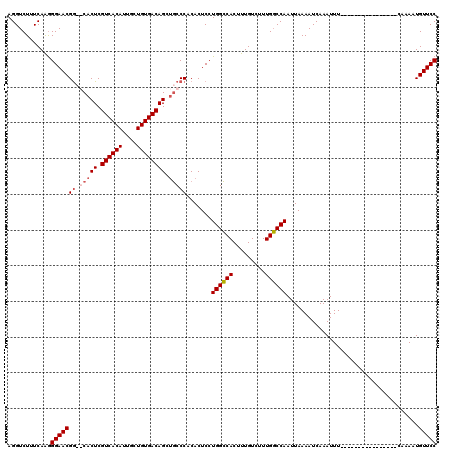
| Location | 3,154,197 – 3,154,307 |
|---|---|
| Length | 110 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 87.61 |
| Mean single sequence MFE | -35.23 |
| Consensus MFE | -30.54 |
| Energy contribution | -29.98 |
| Covariance contribution | -0.56 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.49 |
| Structure conservation index | 0.87 |
| SVM decision value | 1.97 |
| SVM RNA-class probability | 0.984307 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 3154197 110 - 27905053 GGAACAUUUUGA--------AAUUUUGAAAUUUGAUUUUAAUUGGCCAAAGACAAAGUGGCCAGGAGUGUGGGCAGCUGUCACAGCAAUGUGACGAGUG--UCGUUCCCUUGAAAGACCU (((((...((((--------((((.........))))))))(((((((.........))))))).......((((..(((((((....)))))))..))--)))))))............ ( -33.30) >DroSec_CAF1 2085 104 - 1 GGAACAUUUUG----------------AAAUUUGAUUUUAAUUGGCCAAAGACAAAGUGGCCAGGAGUGUGGGCAGCUGUCACAGCAAUGUGACGAGUGCGCCGUUCCCUUGAAAGACCU (((((......----------------..............(((((((.........))))))).......(((.(((((((((....)))))).)))..))))))))............ ( -34.20) >DroSim_CAF1 4255 104 - 1 GGAACAUUUUG----------------AAAUUUGAUUUUAAUUGGCCAAAGACAAAGUGGCCAGGAGUGUGGGCAGCUGUCACAGCAAUGUGACGAGUGCGCCGUUCCCUUGAAAGACCU (((((......----------------..............(((((((.........))))))).......(((.(((((((((....)))))).)))..))))))))............ ( -34.20) >DroYak_CAF1 1383 118 - 1 GGAACAUUUUGAAAUGCUGAAAUUUUGAAAUUUGAUUUUAAUUGGGCAAAGACAAAGUGCCCAGGAGUGUGGGGUGCUGUCACAGCAAUGUGACGAGUG--CCGUUCCUCUGAAAGACCU (((((((((..((((......))))..))))..........(((((((.........)))))))........((..((((((((....)))))).))..--)))))))............ ( -39.20) >consensus GGAACAUUUUG________________AAAUUUGAUUUUAAUUGGCCAAAGACAAAGUGGCCAGGAGUGUGGGCAGCUGUCACAGCAAUGUGACGAGUG__CCGUUCCCUUGAAAGACCU (((((....................................(((((((.........)))))))......((...(((((((((....)))))).)))...)))))))............ (-30.54 = -29.98 + -0.56)

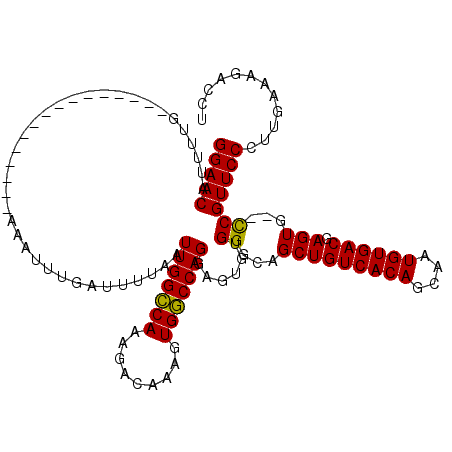
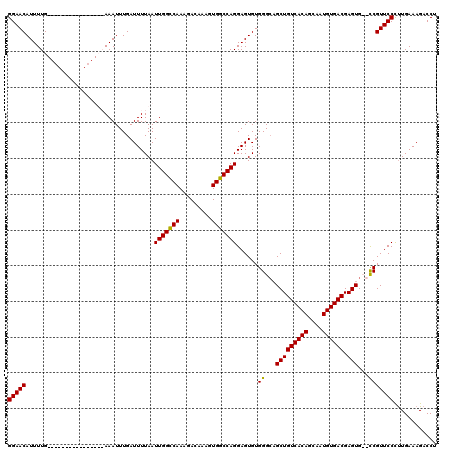
| Location | 3,154,235 – 3,154,346 |
|---|---|
| Length | 111 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 88.65 |
| Mean single sequence MFE | -22.75 |
| Consensus MFE | -19.12 |
| Energy contribution | -19.00 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.11 |
| Structure conservation index | 0.84 |
| SVM decision value | 1.24 |
| SVM RNA-class probability | 0.935271 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 3154235 111 + 27905053 ACAGCUGCCCACACUCCUGGCCACUUUGUCUUUGGCCAAUUAAAAUCAAAUUUCAAAAUU--------UCAAAAUGUUCCAAAUUUUGUGCA-ACUGAUGACAGUUUAAUUGCUGUCCUG .(((.(((.........((((((.........))))))......................--------.((((((.......)))))).)))-.)))..((((((......))))))... ( -24.40) >DroSec_CAF1 2125 104 + 1 ACAGCUGCCCACACUCCUGGCCACUUUGUCUUUGGCCAAUUAAAAUCAAAUUU----------------CAAAAUGUUCCAAAUUUUGUGCAAACUGAUGACAGUUUAAUUGCUGUCCUG .(((.(((.........((((((.........))))))...............----------------((((((.......)))))).)))..)))..((((((......))))))... ( -22.40) >DroSim_CAF1 4295 104 + 1 ACAGCUGCCCACACUCCUGGCCACUUUGUCUUUGGCCAAUUAAAAUCAAAUUU----------------CAAAAUGUUCCAAAUUUUGUGCAAACUGAUGACAGUUUAAUUGCUGUCCUG .(((.(((.........((((((.........))))))...............----------------((((((.......)))))).)))..)))..((((((......))))))... ( -22.40) >DroYak_CAF1 1421 119 + 1 ACAGCACCCCACACUCCUGGGCACUUUGUCUUUGCCCAAUUAAAAUCAAAUUUCAAAAUUUCAGCAUUUCAAAAUGUUCCUAAUUUUGUGCA-ACUGAUGACAGUUUAAUUGCCGUCCUG ...((((..........((((((.........))))))................((((((..((((((....))))))...)))))))))).-...((((.((((...)))).))))... ( -21.80) >consensus ACAGCUGCCCACACUCCUGGCCACUUUGUCUUUGGCCAAUUAAAAUCAAAUUU________________CAAAAUGUUCCAAAUUUUGUGCA_ACUGAUGACAGUUUAAUUGCUGUCCUG .(((.............((((((.........))))))...............................((((((.......))))))......)))..((((((......))))))... (-19.12 = -19.00 + -0.12)

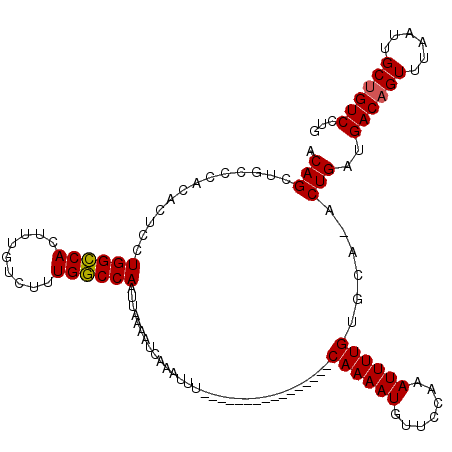
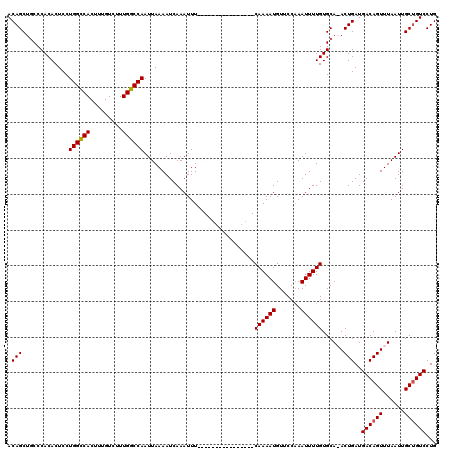
| Location | 3,154,235 – 3,154,346 |
|---|---|
| Length | 111 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.65 |
| Mean single sequence MFE | -30.35 |
| Consensus MFE | -23.58 |
| Energy contribution | -23.77 |
| Covariance contribution | 0.19 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.49 |
| Structure conservation index | 0.78 |
| SVM decision value | 1.32 |
| SVM RNA-class probability | 0.941583 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 3154235 111 - 27905053 CAGGACAGCAAUUAAACUGUCAUCAGU-UGCACAAAAUUUGGAACAUUUUGA--------AAUUUUGAAAUUUGAUUUUAAUUGGCCAAAGACAAAGUGGCCAGGAGUGUGGGCAGCUGU ...(((((........)))))..((((-(((.((((((((.((.....)).)--------)))))))..............(((((((.........)))))))........))))))). ( -31.60) >DroSec_CAF1 2125 104 - 1 CAGGACAGCAAUUAAACUGUCAUCAGUUUGCACAAAAUUUGGAACAUUUUG----------------AAAUUUGAUUUUAAUUGGCCAAAGACAAAGUGGCCAGGAGUGUGGGCAGCUGU ((((((((........)))))....(((..(((.((((((.((.....)).----------------))))))........(((((((.........)))))))..)))..)))..))). ( -29.00) >DroSim_CAF1 4295 104 - 1 CAGGACAGCAAUUAAACUGUCAUCAGUUUGCACAAAAUUUGGAACAUUUUG----------------AAAUUUGAUUUUAAUUGGCCAAAGACAAAGUGGCCAGGAGUGUGGGCAGCUGU ((((((((........)))))....(((..(((.((((((.((.....)).----------------))))))........(((((((.........)))))))..)))..)))..))). ( -29.00) >DroYak_CAF1 1421 119 - 1 CAGGACGGCAAUUAAACUGUCAUCAGU-UGCACAAAAUUAGGAACAUUUUGAAAUGCUGAAAUUUUGAAAUUUGAUUUUAAUUGGGCAAAGACAAAGUGCCCAGGAGUGUGGGGUGCUGU ....((((((....(((((....))))-)((((((((((((....((((..((((......))))..))))))))))))..(((((((.........)))))))..))))....)))))) ( -31.80) >consensus CAGGACAGCAAUUAAACUGUCAUCAGU_UGCACAAAAUUUGGAACAUUUUG________________AAAUUUGAUUUUAAUUGGCCAAAGACAAAGUGGCCAGGAGUGUGGGCAGCUGU ....(((((...(((((((....)))))))((((....((((((((..((..................))..)).))))))(((((((.........)))))))...))))....))))) (-23.58 = -23.77 + 0.19)

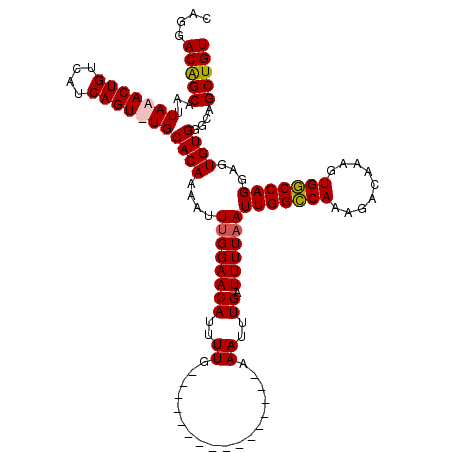
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:56:51 2006