
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 25,521,512 – 25,521,629 |
| Length | 117 |
| Max. P | 0.760010 |

| Location | 25,521,512 – 25,521,629 |
|---|---|
| Length | 117 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 82.23 |
| Mean single sequence MFE | -37.45 |
| Consensus MFE | -26.75 |
| Energy contribution | -26.53 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.26 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.50 |
| SVM RNA-class probability | 0.760010 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
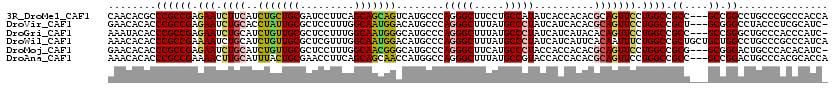
>3R_DroMel_CAF1 25521512 117 + 27905053 CAACACGCCCGCCGAGAAUCUUCAUCUGCUGCGAUCCUUCAGCAGCAGUCAUGCCCAGGGCUUCCUGCCAUAUCACCACACGCAGUUCCUGGCCGCC---GCCGGCCUGCCCGCCCACCA ......((..((.(((....)))..(((((((((....)).)))))))....))...((((...((((.............)))).....(((((..---..))))).))))))...... ( -33.82) >DroVir_CAF1 4144 116 + 1 GAACACACCCGCCGAGAAUCUGCACCUAUUGCGCUCCUUUGGCAAUGGACAUGCCCAGGGCUUUAUGCCCUAUCAUCACACGCAGUUCCUGGCCGCU---GCGGGCCUACCCUCGCAUC- ..........((((.(((.((((..(((((((.(......))))))))........(((((.....)))))..........))))))).))))...(---(((((......))))))..- ( -38.90) >DroGri_CAF1 4744 116 + 1 AAAUACACCCGCCGAGAAUCUGCAUCUGUUGCGCUCCUUUGGCAAUGGGCAUGCCCAGGGCUUUAUGCCCUAUCAUCAUACACAGUUCCUGGCCGCC---GCCGGGCUGCCCACCCAUC- ..........((((((....((((.....))))....))))))..((((((.(((((((((.....)))))...................(((....---)))))))))))))......- ( -43.60) >DroWil_CAF1 12278 120 + 1 AAACACACCCGCCGAAAAUCUGCAUCUGUUGCGCUCGUUUGGCAAUGGACAUGCCCAGGGCUUUAUGCCCUAUCAUCAUUCACAAUUUCUGGCCGCUGCUGCUGGCCUGCCCGCCCAUCA ..........((((((....((((.....))))....)))))).((((.(..((..(((((.....)))))...................((((((....)).)))).))..).)))).. ( -34.60) >DroMoj_CAF1 3484 116 + 1 GAACACACCCGCCGAGAAUCUGCAUCUGUUGCGCUCCUUUGGCAACGGGCAUGCCCAGGGCUUCAUGCCCUACCACCACACGCAGUUCCUGGCCGCG---GCGGGACUGCCCACACAUC- .......(((((((.(((.((((....(((((.(......))))))(((....)))(((((.....)))))..........))))))).......))---)))))..............- ( -42.60) >DroAna_CAF1 2871 117 + 1 AAACACACCCGCCGAAAACUUGCAUUUACUGCGAACCUUCAGCAGCAACCAUGGCCAGGGCUUUAUGCCGUACCACCACACGCAGUUCCUGGCCGCC---GCCGGACUGCCCACGCACCA ..........((.(((...(((((.....)))))...))).)).((.....(((..(.(((.....))).).)))......(((((.((.(((....---))))))))))....)).... ( -31.20) >consensus AAACACACCCGCCGAGAAUCUGCAUCUGUUGCGCUCCUUUGGCAACGGACAUGCCCAGGGCUUUAUGCCCUAUCACCACACGCAGUUCCUGGCCGCC___GCCGGCCUGCCCACCCAUC_ ........((((((.(((.((((..(((((((.........)))))))........(((((.....)))))..........))))))).)))).((....)).))............... (-26.75 = -26.53 + -0.22)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:39:12 2006