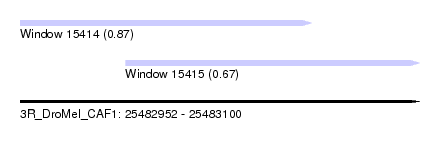
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 25,482,952 – 25,483,100 |
| Length | 148 |
| Max. P | 0.872936 |

| Location | 25,482,952 – 25,483,060 |
|---|---|
| Length | 108 |
| Sequences | 4 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 86.94 |
| Mean single sequence MFE | -39.73 |
| Consensus MFE | -30.51 |
| Energy contribution | -32.20 |
| Covariance contribution | 1.69 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.88 |
| SVM RNA-class probability | 0.872936 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 25482952 108 + 27905053 CAGAAUGCGAAGCCUCA-AACUUGGCUGCAGGCUGCUAUUAUUGUGGCCGCAGCUGUGCACACAGGACUGUGAGCUAGGACCUGUUCUGCUCAGCAACUCGGCGCUGAA (((..((((.((((...-.....)))).).(((..(.......)..))))))((((.(((.(((((.(((.....)))..)))))..))).)))).........))).. ( -39.10) >DroSec_CAF1 164148 108 + 1 CAGAAUGCGAAGCCUCA-AACUUGGCUGCAGGCUGCUAUUAUUGUGGCCGCAGCUGUGCACACAGGACUGUGAGCUAGGACCUGUUCUGCUCAGCUCCUCGGCGCUGAA (((((((.(...(((..-.....((((((.(((..(.......)..)))))))))..((.((((....)))).)).))).).))))))).(((((.(....).))))). ( -40.10) >DroSim_CAF1 166143 108 + 1 CAGAAUGCGAAGCCUAA-AACUUGGCUGCAGGCUGCUAUUAUUGUGGCCGCAGCUGUGCACACAGGACUGUGAGCUAGGACCUGUUCUGCUCAGCUCCUCGGCGCUGAA (((((((.(...((((.-.....((((((.(((..(.......)..)))))))))..((.((((....)))).)))))).).))))))).(((((.(....).))))). ( -42.00) >DroEre_CAF1 171155 91 + 1 CAGAAUGCGAAGCUCCAGCACUUGGCUGCAGGCUGCUAUUAAUGUGGCCGCAGCUGUGCACAUACGACUGUGUGCUAGGACCUGUU------------------CUGAA ((((((....((.(((.((((..((((((.(((..(.......)..))))))))))))).(((((....)))))...))).)))))------------------))).. ( -37.70) >consensus CAGAAUGCGAAGCCUCA_AACUUGGCUGCAGGCUGCUAUUAUUGUGGCCGCAGCUGUGCACACAGGACUGUGAGCUAGGACCUGUUCUGCUCAGCUCCUCGGCGCUGAA (((((((.(...(((........((((((.(((..(.......)..)))))))))..((.((((....)))).)).))).).))))))).(((((........))))). (-30.51 = -32.20 + 1.69)

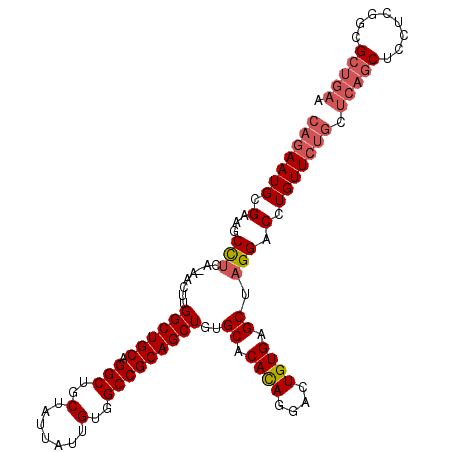
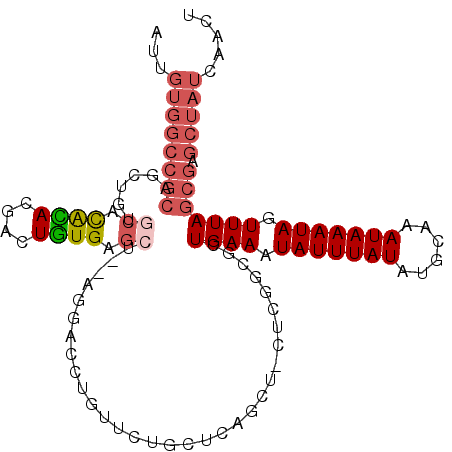
| Location | 25,482,991 – 25,483,100 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 78.00 |
| Mean single sequence MFE | -30.53 |
| Consensus MFE | -12.15 |
| Energy contribution | -13.32 |
| Covariance contribution | 1.17 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.41 |
| Structure conservation index | 0.40 |
| SVM decision value | 0.28 |
| SVM RNA-class probability | 0.666743 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 25482991 109 + 27905053 AUUGUGGCCGCAGCUGUGCACACAGGACUGUGAGCU--AGGACCUGUUCUGCUCAGCAACUCGGCGCUGAAAUAUUUAUAUGCAAAUAAAUAGUUUAGCGAGCUAUCAACU .(((((((....((((.(((.(((((.(((.....)--))..)))))..))).)))).......(((((((.(((((((......))))))).))))))).)))).))).. ( -35.40) >DroSec_CAF1 164187 109 + 1 AUUGUGGCCGCAGCUGUGCACACAGGACUGUGAGCU--AGGACCUGUUCUGCUCAGCUCCUCGGCGCUGAAAUAUUUAUAUGCAAAUAAAUAGUUUAGCGAGCUAUCAACU ......((((.(((((.(((.(((((.(((.....)--))..)))))..))).)))))...))))((((((.(((((((......))))))).))))))............ ( -36.70) >DroSim_CAF1 166182 109 + 1 AUUGUGGCCGCAGCUGUGCACACAGGACUGUGAGCU--AGGACCUGUUCUGCUCAGCUCCUCGGCGCUGAAAUAUUUAUAUGCAAAUAAAUAGUUUAGCGAGCUAUCAACU ......((((.(((((.(((.(((((.(((.....)--))..)))))..))).)))))...))))((((((.(((((((......))))))).))))))............ ( -36.70) >DroEre_CAF1 171195 91 + 1 AAUGUGGCCGCAGCUGUGCACAUACGACUGUGUGCU--AGGACCUGUU------------------CUGAAAUAUUUAUAUGCAAAUAAAUAGUUUAGCGAGCUAUCAACU ...((((((((..(((.(((((((....))))))))--))........------------------.((((.(((((((......))))))).))))))).)))))..... ( -23.60) >DroYak_CAF1 177432 95 + 1 AUUGUGGCCGCAGCUGUUCACAUACGACUGUGUGCUAUAGGACCUGUUCU----------------CUGAAAUAUUUAUAUGCAAAUAAAUAGUUUAGCGAGCUAUCAGCU .((((((((((((.(((......))).))))).)))))))((...((((.----------------(((((.(((((((......))))))).))))).))))..)).... ( -28.20) >DroAna_CAF1 173436 88 + 1 -------------------AUGUACGAGUAUGUGCU--AAGAGCAGUG--GGUCCUUUGCUCGCUUCUGAAAUAUUUAUAUGCAAAUAAAUAGUUUAGCGAGCUAUCAACU -------------------..((((......)))).--..((((((.(--....).))))))(((((((((.(((((((......))))))).))))).))))........ ( -22.60) >consensus AUUGUGGCCGCAGCUGUGCACACACGACUGUGAGCU__AGGACCUGUUCUGCUCAGCU_CUCGGCGCUGAAAUAUUUAUAUGCAAAUAAAUAGUUUAGCGAGCUAUCAACU ...((((((((......((.((((....)))).))................................((((.(((((((......))))))).))))))).)))))..... (-12.15 = -13.32 + 1.17)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:38:57 2006