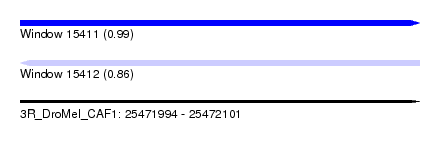
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 25,471,994 – 25,472,101 |
| Length | 107 |
| Max. P | 0.988138 |

| Location | 25,471,994 – 25,472,101 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 77.52 |
| Mean single sequence MFE | -29.25 |
| Consensus MFE | -19.84 |
| Energy contribution | -21.53 |
| Covariance contribution | 1.70 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.90 |
| Structure conservation index | 0.68 |
| SVM decision value | 2.11 |
| SVM RNA-class probability | 0.988138 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
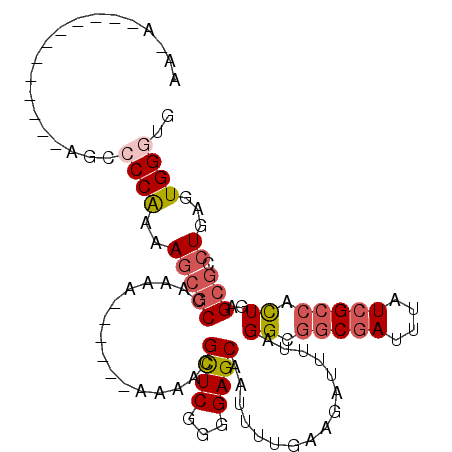
>3R_DroMel_CAF1 25471994 107 + 27905053 CACCCACUCAGGCGCUCAGUGGCGAUAAAUCGCCGCCUAAAAUCUUCAAAAUUGCUCUGCGAGCGUUUUUUUUUUUUUUGGCGCUUUUGGGGCUUCGAUCCU--UUU-UU ..((((...((((((...(((((((....))))))).(((((.....((((.(((((...))))).)))).....))))))))))).))))...........--...-.. ( -31.50) >DroSec_CAF1 154880 103 + 1 CACCCACUCAGGCGCUCAGUGGCCAUAAAUCGCCGCCUAAAAUCUUCAAAAUUGCUCCCCGAGCGAUUU------UUUUGGCGCUUUUGGGGCUUCGCUCCAUUUUU-UU .........((((((.(((((((........)))))...........((((((((((...)))))))))------)..)))))))).((((((...)))))).....-.. ( -31.80) >DroSim_CAF1 156922 104 + 1 CACCCACUCAGCCGCUCAGUGGCGAUAAAUCGCCGCCUAAAAUCUUCAAAAUUGCUCCCCGAGCGAUUU------UUUUGGCGCUUUUGGGGCUUCGCUCCAUUUUUUUU ..........(((.....(((((((....)))))))...........((((((((((...)))))))))------)...))).....((((((...))))))........ ( -32.40) >DroEre_CAF1 162133 88 + 1 CAGCCACUCAGCCGCUCAGCGGCGAUAAAUCGCCGCCUAAAGUCUUCAGAAUUGCUCCCCGAGCGUUU--------UUUGGCGCUU-UGGGGCU------------U-UU .........((((.....(((((((....)))))))(((((((..((((((.(((((...)))))..)--------))))).))))-)))))))------------.-.. ( -33.50) >DroYak_CAF1 167136 96 + 1 CACCCACUUAGGCGCUCAAUGGCGAUAAAUCGCCGCCUAAAAUCUUCCAAAUUGCUCCCUGAACAUUUUUU-UUUUUUUGGCGCUUUUGGGGCU------------U-UU ..((((...((((((.(((.(((((....)))))....(((((.(((.............))).)))))..-.....))))))))).))))...------------.-.. ( -24.32) >DroAna_CAF1 164026 82 + 1 CACCCACUCAGGAGCUGAG---CGAUAAAUCGCUGCCUAAAAUCUUCCAAAUUGCUCCCAGAGCGUUUU---------CAGGGUUUUCGG--C-------------A-UU ...((.....)).....((---(((....)))))(((.((((((((..((((.((((...)))))))).---------.)))))))).))--)-------------.-.. ( -22.00) >consensus CACCCACUCAGGCGCUCAGUGGCGAUAAAUCGCCGCCUAAAAUCUUCAAAAUUGCUCCCCGAGCGUUUU______UUUUGGCGCUUUUGGGGCU____________U_UU ..((((...(((((((..(((((((....)))))))................(((((...)))))..............))))))).))))................... (-19.84 = -21.53 + 1.70)

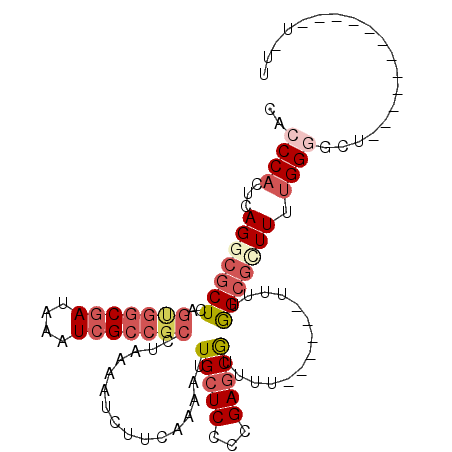
| Location | 25,471,994 – 25,472,101 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 77.52 |
| Mean single sequence MFE | -28.27 |
| Consensus MFE | -17.36 |
| Energy contribution | -18.28 |
| Covariance contribution | 0.92 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.81 |
| SVM RNA-class probability | 0.856240 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 25471994 107 - 27905053 AA-AAA--AGGAUCGAAGCCCCAAAAGCGCCAAAAAAAAAAAAAACGCUCGCAGAGCAAUUUUGAAGAUUUUAGGCGGCGAUUUAUCGCCACUGAGCGCCUGAGUGGGUG ..-...--.........(((((..(.((((................((((...)))).............((((..(((((....))))).)))))))).)..).)))). ( -27.30) >DroSec_CAF1 154880 103 - 1 AA-AAAAAUGGAGCGAAGCCCCAAAAGCGCCAAAA------AAAUCGCUCGGGGAGCAAUUUUGAAGAUUUUAGGCGGCGAUUUAUGGCCACUGAGCGCCUGAGUGGGUG ..-.....(((.((............)).)))...------...(((((((((..((((((.....))))((((..(((........))).)))))).)))))))))... ( -27.40) >DroSim_CAF1 156922 104 - 1 AAAAAAAAUGGAGCGAAGCCCCAAAAGCGCCAAAA------AAAUCGCUCGGGGAGCAAUUUUGAAGAUUUUAGGCGGCGAUUUAUCGCCACUGAGCGGCUGAGUGGGUG ........(((.((...)).)))....((((....------.(..(((((((...((.................))(((((....))))).)))))))..).....)))) ( -29.43) >DroEre_CAF1 162133 88 - 1 AA-A------------AGCCCCA-AAGCGCCAAA--------AAACGCUCGGGGAGCAAUUCUGAAGACUUUAGGCGGCGAUUUAUCGCCGCUGAGCGGCUGAGUGGCUG ..-.------------..((((.-.((((.....--------...)))).))))(((.((((.(..(.(((..((((((((....))))))))))))..).)))).))). ( -34.30) >DroYak_CAF1 167136 96 - 1 AA-A------------AGCCCCAAAAGCGCCAAAAAAA-AAAAAAUGUUCAGGGAGCAAUUUGGAAGAUUUUAGGCGGCGAUUUAUCGCCAUUGAGCGCCUAAGUGGGUG ..-.------------.(((((....(((((((.....-..(((((.(((..(((....))).))).)))))....(((((....))))).))).))))....).)))). ( -27.90) >DroAna_CAF1 164026 82 - 1 AA-U-------------G--CCGAAAACCCUG---------AAAACGCUCUGGGAGCAAUUUGGAAGAUUUUAGGCAGCGAUUUAUCG---CUCAGCUCCUGAGUGGGUG ..-.-------------.--......((((..---------.....((((.((((((...(((((....)))))(.(((((....)))---))).)))))))))))))). ( -23.31) >consensus AA_A____________AGCCCCAAAAGCGCCAAAA______AAAACGCUCGGGGAGCAAUUUUGAAGAUUUUAGGCGGCGAUUUAUCGCCACUGAGCGCCUGAGUGGGUG ...................((((..(((((................((((...))))................((.(((((....))))).))..))).))...)))).. (-17.36 = -18.28 + 0.92)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:38:55 2006