
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 25,464,715 – 25,464,807 |
| Length | 92 |
| Max. P | 0.942986 |

| Location | 25,464,715 – 25,464,807 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 76.31 |
| Mean single sequence MFE | -30.45 |
| Consensus MFE | -18.23 |
| Energy contribution | -18.90 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.48 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.24 |
| SVM RNA-class probability | 0.650940 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
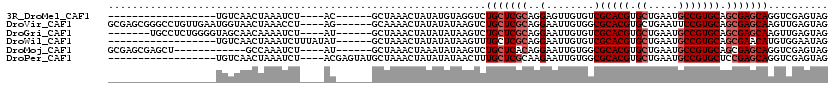
>3R_DroMel_CAF1 25464715 92 + 27905053 ------------------UGUCAACUAAAUCU----AC------GCUAAACUAUAUGUAGGUCUGCUCGCAGGAGUUGUGUCGCACGUGCUGAAUGCCGUGCAGCGAGCAGGUCGAGUAG ------------------............((----((------.....((.....)).(..((((((((..((......))(((((.((.....))))))).))))))))..)..)))) ( -33.60) >DroVir_CAF1 177085 110 + 1 GCGAGCGGGCCUGUUGAAUGGUAACUAAACCU----AG------GCAAAACUAUAUAUAAGUCUGCUCGCAGGAAUUGUGGCGCACGUGCUGAAUUCCGUGCAGCGAGCAAGUUGAGUAG ((((((((((.(((...(((((..(((....)----))------.....)))))..))).)))))))))).((((((.(((((....)))))))))))...((((......))))..... ( -36.40) >DroGri_CAF1 173975 103 + 1 -------UGCCUCUGGGGUAGCAACAAAAUCU----AU------GCUAAACUAUAUAUAAGUCUGCUCGCAGGAAUUGUGUCGCACGUGCUGAAUGCCGUGCAGCGAGCAAGUUGAGUAG -------...((((((..(((((.........----.)------))))..)))......((..(((((((..((......))(((((.((.....))))))).)))))))..)))))... ( -33.50) >DroWil_CAF1 183816 96 + 1 ------------------UGUCAACUAAAUCUUUAUAU------GCUAAACUAUAUAUAAGUUUGCUCGCAGGAAUUGUGUCGCACGUGCUGAAUGCCGUGCAGCGAACAUGUGGAAUAG ------------------....................------......((((.((((.((((((((((((...)))))..(((((.((.....)))))))))))))).))))..)))) ( -23.90) >DroMoj_CAF1 175109 98 + 1 GCGAGCGAGCU------------GCCAAAUCU----AU------GCUAAACUAAAUAUAAGUCUGCUCACAGGAAUUGUGGCGCACGUGCUGAAUGCCGUGCAGCGAGCAGGUCGAGUAG ((...(((.((------------(((......----..------....................((.(((((...)))))))(((((.((.....)))))))...).)))).))).)).. ( -29.30) >DroPer_CAF1 158184 98 + 1 ------------------UGUCAACUAAAUCU----ACGAGUAUGCUAAACUAUAUAUAACUUUGCUCGCAAGAAUUGUGGCGCACGUGCUGAAUGCCGUGCUCCGAGCAGGUCGAGUAG ------------------............((----((..(((((......))))).....((((((((((.....)))((.(((((.((.....))))))).)))))))))....)))) ( -26.00) >consensus __________________UGUCAACUAAAUCU____AU______GCUAAACUAUAUAUAAGUCUGCUCGCAGGAAUUGUGGCGCACGUGCUGAAUGCCGUGCAGCGAGCAGGUCGAGUAG ...............................................................(((((((..(........)(((((.((.....))))))).))))))).......... (-18.23 = -18.90 + 0.67)


| Location | 25,464,715 – 25,464,807 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 76.31 |
| Mean single sequence MFE | -27.17 |
| Consensus MFE | -19.07 |
| Energy contribution | -19.52 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.70 |
| SVM decision value | 1.33 |
| SVM RNA-class probability | 0.942986 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 25464715 92 - 27905053 CUACUCGACCUGCUCGCUGCACGGCAUUCAGCACGUGCGACACAACUCCUGCGAGCAGACCUACAUAUAGUUUAGC------GU----AGAUUUAGUUGACA------------------ ....(((((((((((((.(((((((.....)).)))))((......))..))))))))..((((.((.....))..------))----)).....)))))..------------------ ( -31.00) >DroVir_CAF1 177085 110 - 1 CUACUCAACUUGCUCGCUGCACGGAAUUCAGCACGUGCGCCACAAUUCCUGCGAGCAGACUUAUAUAUAGUUUUGC------CU----AGGUUUAGUUACCAUUCAACAGGCCCGCUCGC ...............((.(((((..........)))))))..........(((((((((((.......))))).((------((----.(((......))).......))))..)))))) ( -25.70) >DroGri_CAF1 173975 103 - 1 CUACUCAACUUGCUCGCUGCACGGCAUUCAGCACGUGCGACACAAUUCCUGCGAGCAGACUUAUAUAUAGUUUAGC------AU----AGAUUUUGUUGCUACCCCAGAGGCA------- ...(((...((((((((.(((((((.....)).)))))((......))..))))))))...........(..((((------(.----.........)))))..)..)))...------- ( -29.80) >DroWil_CAF1 183816 96 - 1 CUAUUCCACAUGUUCGCUGCACGGCAUUCAGCACGUGCGACACAAUUCCUGCGAGCAAACUUAUAUAUAGUUUAGC------AUAUAAAGAUUUAGUUGACA------------------ ....((.((.(((((((.(((((((.....)).)))))((......))..)))))))..(((.(((((.(.....)------)))))))).....)).))..------------------ ( -21.90) >DroMoj_CAF1 175109 98 - 1 CUACUCGACCUGCUCGCUGCACGGCAUUCAGCACGUGCGCCACAAUUCCUGUGAGCAGACUUAUAUUUAGUUUAGC------AU----AGAUUUGGC------------AGCUCGCUCGC .....(((.((((..((.(((((((.....)).)))))))..(((...(((((...(((((.......)))))..)------))----))..)))))------------)).)))..... ( -28.70) >DroPer_CAF1 158184 98 - 1 CUACUCGACCUGCUCGGAGCACGGCAUUCAGCACGUGCGCCACAAUUCUUGCGAGCAAAGUUAUAUAUAGUUUAGCAUACUCGU----AGAUUUAGUUGACA------------------ ....(((((..(.((((.(((((((.....)).))))).))........((((((....((((.........))))...)))))----))).)..)))))..------------------ ( -25.90) >consensus CUACUCGACCUGCUCGCUGCACGGCAUUCAGCACGUGCGACACAAUUCCUGCGAGCAGACUUAUAUAUAGUUUAGC______AU____AGAUUUAGUUGACA__________________ .........((((((((((((((((.....)).))))))...........)))))))).............................................................. (-19.07 = -19.52 + 0.45)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:38:51 2006