
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 25,462,346 – 25,462,441 |
| Length | 95 |
| Max. P | 0.725894 |

| Location | 25,462,346 – 25,462,441 |
|---|---|
| Length | 95 |
| Sequences | 4 |
| Columns | 95 |
| Reading direction | forward |
| Mean pairwise identity | 84.93 |
| Mean single sequence MFE | -14.90 |
| Consensus MFE | -9.46 |
| Energy contribution | -9.96 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.09 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.41 |
| SVM RNA-class probability | 0.725894 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
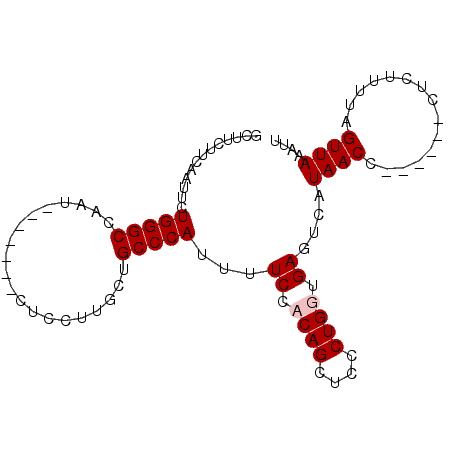
>3R_DroMel_CAF1 25462346 95 + 27905053 GCUUUUUCAAUUCUGGGCCAAUCUCCAAUCUCCUUGCUGCCCAUUUUCCCCAGCUCGCUGGUGAGUCAUAACCUUUAACCUCUUUUGGUUAAAUU .............((((((((............)))..)))))..(((.((((....)))).)))........(((((((......))))))).. ( -19.50) >DroSec_CAF1 145394 82 + 1 GCUUUUUCAAUUCUGGGCCAAU-------CUCCUUGCUGCCCAUUUUCCCCAGCUCGCUGGUGAUUCAUAACC------CUCUUUUGGUUAAAUU .............((((((((.-------....)))..)))))...((.((((....)))).))....(((((------.......))))).... ( -16.90) >DroEre_CAF1 152712 80 + 1 UCUUCUUCAAUUCUGGGCUAAU-------CUCCCUACUGCCCAUU-UCCACAGCUCCCUGGUGAGUCAUAACC------CUCU-UUAGUUAAAUU .............(((((....-------.........)))))..-...........((((.(((........------))).-))))....... ( -10.82) >DroYak_CAF1 155908 82 + 1 UCUUCUUCAAUUCUGGGCCAAU-------CUCCUUGCUGCCCAUUUUCCACAGCUCCCUGGUGAGUCAUAACC------CACUUUUAGUUAAAUU .............((((((((.-------....)))..)))))........((((....((((.((....)).------))))...))))..... ( -12.40) >consensus GCUUCUUCAAUUCUGGGCCAAU_______CUCCUUGCUGCCCAUUUUCCACAGCUCCCUGGUGAGUCAUAACC______CUCUUUUAGUUAAAUU .............(((((....................)))))...((.((((....)))).))....((((...............)))).... ( -9.46 = -9.96 + 0.50)


| Location | 25,462,346 – 25,462,441 |
|---|---|
| Length | 95 |
| Sequences | 4 |
| Columns | 95 |
| Reading direction | reverse |
| Mean pairwise identity | 84.93 |
| Mean single sequence MFE | -18.50 |
| Consensus MFE | -13.18 |
| Energy contribution | -12.87 |
| Covariance contribution | -0.31 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.68 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.29 |
| SVM RNA-class probability | 0.671925 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 25462346 95 - 27905053 AAUUUAACCAAAAGAGGUUAAAGGUUAUGACUCACCAGCGAGCUGGGGAAAAUGGGCAGCAAGGAGAUUGGAGAUUGGCCCAGAAUUGAAAAAGC ..(((((((......))))))).........((.((((....)))).))...(((((..(((.....))).......)))))............. ( -20.30) >DroSec_CAF1 145394 82 - 1 AAUUUAACCAAAAGAG------GGUUAUGAAUCACCAGCGAGCUGGGGAAAAUGGGCAGCAAGGAG-------AUUGGCCCAGAAUUGAAAAAGC ....(((((.......------)))))....((.((((....)))).))...(((((..(((....-------.))))))))............. ( -18.80) >DroEre_CAF1 152712 80 - 1 AAUUUAACUAA-AGAG------GGUUAUGACUCACCAGGGAGCUGUGGA-AAUGGGCAGUAGGGAG-------AUUAGCCCAGAAUUGAAGAAGA ...........-...(------(((((...(((.((.....(((((...-.....))))).)))))-------..)))))).............. ( -17.10) >DroYak_CAF1 155908 82 - 1 AAUUUAACUAAAAGUG------GGUUAUGACUCACCAGGGAGCUGUGGAAAAUGGGCAGCAAGGAG-------AUUGGCCCAGAAUUGAAGAAGA ..((((((((...(((------(((....))))))(((....))))))....(((((..(((....-------.))))))))...)))))..... ( -17.80) >consensus AAUUUAACCAAAAGAG______GGUUAUGACUCACCAGCGAGCUGGGGAAAAUGGGCAGCAAGGAG_______AUUGGCCCAGAAUUGAAAAAGA ....(((((.............)))))....((.((((....)))).))...(((((..(((............))))))))............. (-13.18 = -12.87 + -0.31)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:38:48 2006