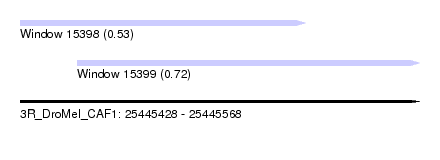
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 25,445,428 – 25,445,568 |
| Length | 140 |
| Max. P | 0.723152 |

| Location | 25,445,428 – 25,445,528 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 85.80 |
| Mean single sequence MFE | -34.75 |
| Consensus MFE | -26.48 |
| Energy contribution | -26.98 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.20 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.76 |
| SVM decision value | -0.00 |
| SVM RNA-class probability | 0.532079 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 25445428 100 + 27905053 AUCCCAAAUCCGCAGGUCAAGACCCGCAUGCAGAUUUCUGGAGCGGGCAGUGGCAAGAAGGAGUACCGCAGCUCAUUGCACUGCAUCCAGACCAUCGUGA ..........(((.((((.....((((.(.(((....))).)))))((((((.(((....((((......)))).))))))))).....))))...))). ( -35.00) >DroSec_CAF1 128349 100 + 1 AUCCCUAAUCCGCAGGUCAAGACCCGCAUGCAGAUUUCUGGAGCAGGCAGUGGCAAGAAGGAGUACCGCAGCUCCUUGCACUGCAUCCAGACCAUCGUGA ...........((.(((....))).)).....(((.((((((....((((((.((...((((((......)))))))))))))).))))))..))).... ( -35.20) >DroSim_CAF1 130196 100 + 1 AUCCCCAAUCCGCAGGUCAAGACCCGCAUGCAGAUUUCUGGAGCGGGCAGUGGCAAGAAGGAGUACCGCAGCUCCUUGCACUGCAUCCAGACCAUCGUGA ..........(((.((((.....((((.(.(((....))).)))))((((((.((...((((((......)))))))))))))).....))))...))). ( -37.10) >DroEre_CAF1 134720 100 + 1 AACUUUGAUUCGCAGGUCAAGACCCGCAUGCAAAUCUCUGGAGCCGGCGGCGGCAAGAAGGAGUACCGCAACUCCUUGCACUGCAUCCAGACCAUCAUGA .....((((..((.(((....))).)).........((((((....((((..(((...((((((......))))))))).)))).))))))..))))... ( -31.70) >DroYak_CAF1 138776 100 + 1 AUUAUUGAUUCACAGGUCAAGACGCGCAUGCAAAUCUCUGGAGCCGGCAGUGGCAAGAAGGAGUACCGCAGCUCCUUGCACUGCAUCCAGACCAUCGUGA ....(((((......)))))..((((.(((......((((((....((((((.((...((((((......)))))))))))))).)))))).))))))). ( -34.30) >DroAna_CAF1 136795 100 + 1 CUCUCUCUUCGAUAGGUGAAAACCCGCAUGCAGAUCUCUGGAGCUGGCAGUGGAAAGAAGGAGUUCAGGAACUCCUUCCAUUGCAUGCAAACGAUUGUCA ..........(((((((....))).((((.(((.((....)).)))((((((((....(((((((....))))))))))))))))))).......)))). ( -35.20) >consensus AUCCCUAAUCCGCAGGUCAAGACCCGCAUGCAGAUCUCUGGAGCCGGCAGUGGCAAGAAGGAGUACCGCAGCUCCUUGCACUGCAUCCAGACCAUCGUGA ......((((.((((((....)))....))).))))((((((....((((((.((...((((((......)))))))))))))).))))))......... (-26.48 = -26.98 + 0.50)


| Location | 25,445,448 – 25,445,568 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 75.28 |
| Mean single sequence MFE | -45.52 |
| Consensus MFE | -22.12 |
| Energy contribution | -23.10 |
| Covariance contribution | 0.98 |
| Combinations/Pair | 1.36 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.49 |
| SVM decision value | 0.40 |
| SVM RNA-class probability | 0.723152 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 25445448 120 + 27905053 GACCCGCAUGCAGAUUUCUGGAGCGGGCAGUGGCAAGAAGGAGUACCGCAGCUCAUUGCACUGCAUCCAGACCAUCGUGAGCAAGGAAGGACCUCUGGCCCUUUACCAGGGCAUCGGAGC ...(((....((((..((((((((((((((((((.....((....))...)).)))))).)))).))))))((.((.(.....).)).))...))))(((((.....)))))..)))... ( -44.30) >DroGri_CAF1 155226 120 + 1 GAAUCGAAUGCAGAUAGCUGGUGCCGGUAGCGGCAAGAAGGAAUACCGCAAUUCCUUCCACUGCAUUCAGACGGUGGUUAAGCGCGAGGGACCCCUUGCCCUGUACCAGGGCAUUUCAGC (((..((((((((........(((((....))))).((((((((......))))))))..))))))))....((.((((...(....).)))))).(((((((...))))))).)))... ( -48.90) >DroWil_CAF1 162576 120 + 1 GACCCGCAUGCAAAUUUCUGGUGCCGGUAGUGGCAAAAAAGAAUUCCGUAAUUCGUUCCACUGCAUACAGACUGUGAUCUGUCGGGAAGGACCGUUGGCUCUGUACCAGGGUAUUGGAGC ..((((((..((....((((......(((((((.((....(((((....))))).)))))))))...))))...))...)).)))).....(((...((((((...))))))..)))... ( -34.00) >DroMoj_CAF1 153231 120 + 1 GAAUCGAAUGCAAAUCGCUGGAGCGGGUAGCGGCAAAAAGGAAUUUCGUAACUCCUUCCACUGCAUUCAGACGGUGAUCAGCCGCGAAGGACCCCUCGCCCUCUAUCAGGGUAUUUCAGC ................(((((((.((((.(((((...(((((.((....)).))))).(((((........)))))....))))).....)))))))(((((.....)))))....)))) ( -40.40) >DroAna_CAF1 136815 120 + 1 AACCCGCAUGCAGAUCUCUGGAGCUGGCAGUGGAAAGAAGGAGUUCAGGAACUCCUUCCAUUGCAUGCAAACGAUUGUCAGCAAGGAGGGAGUCCUGGCUCUCUACACGGGAAUCGGAGC ..(((((.(.(((....))).)....)).(((...(((..((((.(((((.((((((((.((((..((((....))))..)))))))))))))))))))))))).))))))......... ( -47.80) >DroPer_CAF1 138539 120 + 1 GACACGGAUGCAGAUUUCUGGGGCCGGAAGCGGCAAGAAGGAGUUCCGCAACUCCUUCCACUGCAUCCAGCAGAUAGUCGCCCGGGAGGGGCCCCUGUCUCUCUACCAGGGCAUUGGUGC .....((((((((.........((((....))))..(((((((((....)))))))))..)))))))).(((..((((.((((.((((((((....))))))))....)))))))).))) ( -57.70) >consensus GACCCGCAUGCAGAUUUCUGGAGCCGGUAGCGGCAAGAAGGAAUUCCGCAACUCCUUCCACUGCAUCCAGACGAUGGUCAGCCGGGAAGGACCCCUGGCCCUCUACCAGGGCAUUGGAGC .....((((((((.........((((....))))...((((((((....))))))))...)))))))).............................(((((.....)))))........ (-22.12 = -23.10 + 0.98)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:38:44 2006