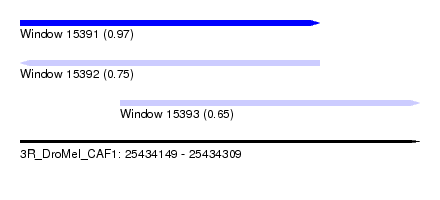
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 25,434,149 – 25,434,309 |
| Length | 160 |
| Max. P | 0.969071 |

| Location | 25,434,149 – 25,434,269 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 88.75 |
| Mean single sequence MFE | -38.64 |
| Consensus MFE | -31.66 |
| Energy contribution | -33.34 |
| Covariance contribution | 1.69 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.51 |
| Structure conservation index | 0.82 |
| SVM decision value | 1.64 |
| SVM RNA-class probability | 0.969071 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
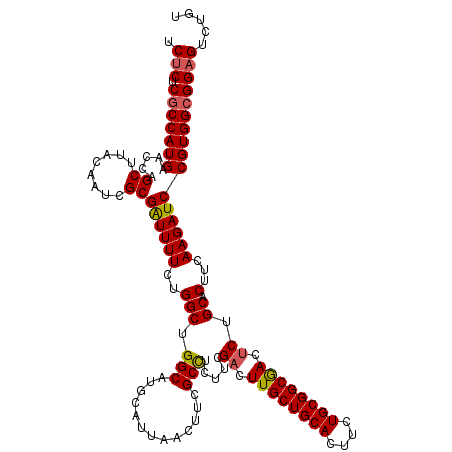
>3R_DroMel_CAF1 25434149 120 + 27905053 UAGCAGUGCAGCAGGAGAAGAUUAUUCAGAUUCCUCUGAAUCUCUCGCCAUGAACCAGCUAACAAUCGCGAUUUUCUGGCUGGCAUGCAUUAACUUCGCCUCUUCGACUUGCUGCACUUU ....(((((((((((.(((((..(((((((....))))))).....((.(((..(((((((..(((....)))...))))))))))))............)))))..))))))))))).. ( -41.80) >DroSec_CAF1 116985 119 + 1 UAGCA-UGCAGCUGGAGAAGAUAAUUCAGAUUCCCGUGGAUCUCUCGCCAUGAAGCAGCUUACAAUCGCGAUUUUCUGGCUGGCAUGCAUUAACUUCGCCUCUUCGACUUGCUGCACUUC .....-((((((.(..(((((......((((((....))))))...((((.((((..((........))...)))))))).(((.............))))))))..)..)))))).... ( -32.82) >DroSim_CAF1 118915 120 + 1 UAGCAGUGCAGCAGGAGAAGAUUAUUCAGAUUCCUCUAGAUCUCUCGCCAUGAACCGGCUCACAACCGCGAUUUUCUGGCUGGCAUGCAUUAACUUCGCCUCUUCGACUUGCUGCACUUC ....(((((((((((.(((((......(((((......)))))...(((.......)))........((((.......((......)).......)))).)))))..))))))))))).. ( -36.84) >DroYak_CAF1 127231 120 + 1 UAGCAGUGCAGUAGGAGAAGACGAAUCAGAUGUCUCUGGAUCGCUCGCCAUGAAGCGGCUUACAAUCGCGGUUUUCUGGCUGGCAUGUAUAAACUUGGCUUCCACGACUUGCUGCACUUC ....(((((((((((.((((.(((.(((((....))))).))(((.((((.(((((.((........)).))))).)))).))).............))))).....))))))))))).. ( -43.10) >consensus UAGCAGUGCAGCAGGAGAAGAUUAUUCAGAUUCCUCUGGAUCUCUCGCCAUGAACCAGCUUACAAUCGCGAUUUUCUGGCUGGCAUGCAUUAACUUCGCCUCUUCGACUUGCUGCACUUC ....(((((((((((.(((((..((((((......)))))).....((((.((((..((........))...)))))))).(((.............))))))))..))))))))))).. (-31.66 = -33.34 + 1.69)


| Location | 25,434,149 – 25,434,269 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.75 |
| Mean single sequence MFE | -37.95 |
| Consensus MFE | -30.10 |
| Energy contribution | -30.85 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.47 |
| SVM RNA-class probability | 0.749330 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 25434149 120 - 27905053 AAAGUGCAGCAAGUCGAAGAGGCGAAGUUAAUGCAUGCCAGCCAGAAAAUCGCGAUUGUUAGCUGGUUCAUGGCGAGAGAUUCAGAGGAAUCUGAAUAAUCUUCUCCUGCUGCACUGCUA ..(((((((((.(..(((((((((..((....)).)))).((((...((((((........)).))))..)))).....(((((((....)))))))..)))))..)))))))))).... ( -43.60) >DroSec_CAF1 116985 119 - 1 GAAGUGCAGCAAGUCGAAGAGGCGAAGUUAAUGCAUGCCAGCCAGAAAAUCGCGAUUGUAAGCUGCUUCAUGGCGAGAGAUCCACGGGAAUCUGAAUUAUCUUCUCCAGCUGCA-UGCUA ...(((((((..(..(((((((((..((....)).))))...((((...(((.((((....(((.......)))....))))..)))...)))).....)))))..).))))))-).... ( -32.40) >DroSim_CAF1 118915 120 - 1 GAAGUGCAGCAAGUCGAAGAGGCGAAGUUAAUGCAUGCCAGCCAGAAAAUCGCGGUUGUGAGCCGGUUCAUGGCGAGAGAUCUAGAGGAAUCUGAAUAAUCUUCUCCUGCUGCACUGCUA ..(((((((((.(..(((((((((..((....)).))))...((((...((..((((....((((.....))))....))))..))....)))).....)))))..)))))))))).... ( -39.30) >DroYak_CAF1 127231 120 - 1 GAAGUGCAGCAAGUCGUGGAAGCCAAGUUUAUACAUGCCAGCCAGAAAACCGCGAUUGUAAGCCGCUUCAUGGCGAGCGAUCCAGAGACAUCUGAUUCGUCUUCUCCUACUGCACUGCUA ..(((((((...((((((.((((...(((((((..(((.............)))..))))))).))))))))))((((((..((((....))))..))).)))......))))))).... ( -36.52) >consensus GAAGUGCAGCAAGUCGAAGAGGCGAAGUUAAUGCAUGCCAGCCAGAAAAUCGCGAUUGUAAGCCGCUUCAUGGCGAGAGAUCCAGAGGAAUCUGAAUAAUCUUCUCCUGCUGCACUGCUA ..(((((((((.(..((((((((...((....))..))).(((((((....((........))...))).))))........((((....)))).....)))))..)))))))))).... (-30.10 = -30.85 + 0.75)

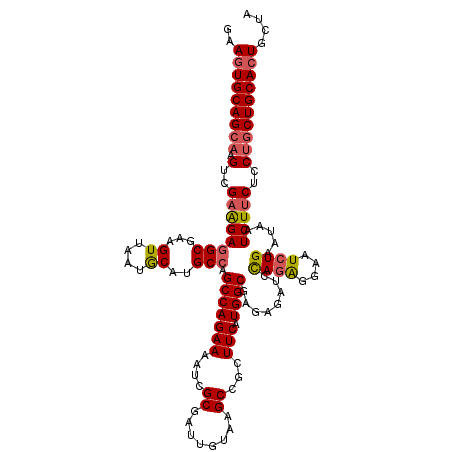
| Location | 25,434,189 – 25,434,309 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.94 |
| Mean single sequence MFE | -39.37 |
| Consensus MFE | -31.51 |
| Energy contribution | -31.70 |
| Covariance contribution | 0.19 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.24 |
| SVM RNA-class probability | 0.648422 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 25434189 120 + 27905053 UCUCUCGCCAUGAACCAGCUAACAAUCGCGAUUUUCUGGCUGGCAUGCAUUAACUUCGCCUCUUCGACUUGCUGCACUUUUGCGGCGACUCUGCACUUCAAGAUCCGUGGCGGAGUCUGU .(((.(((((((..(((((((..(((....)))...)))))))..((((......(((......))).((((((((....))))))))...))))..........))))))))))..... ( -37.70) >DroSec_CAF1 117024 120 + 1 UCUCUCGCCAUGAAGCAGCUUACAAUCGCGAUUUUCUGGCUGGCAUGCAUUAACUUCGCCUCUUCGACUUGCUGCACUUCUGCGGCAACGCUGCACUUCAAGAUCCGUGGCGGAGUCAGU .(((.(((((((..(((((........((((.......((......)).......)))).........((((((((....)))))))).))))).(.....)...))))))))))..... ( -40.64) >DroSim_CAF1 118955 120 + 1 UCUCUCGCCAUGAACCGGCUCACAACCGCGAUUUUCUGGCUGGCAUGCAUUAACUUCGCCUCUUCGACUUGCUGCACUUCUGCGGCGACUCUGCACUUCAAGAUCCGUGGCGGAGUCUGU .(((.(((((((...(((.......))).((((((..(((.(((.............))).....((.((((((((....)))))))).)).)).)...))))))))))))))))..... ( -36.22) >DroYak_CAF1 127271 120 + 1 UCGCUCGCCAUGAAGCGGCUUACAAUCGCGGUUUUCUGGCUGGCAUGUAUAAACUUGGCUUCCACGACUUGCUGCACUUCUGCGGCGACUCUGCACUUCAAGAUCCGUGGAGGAGUCUGC ..(((.((((.(((((.((........)).))))).)))).)))..............((((((((..((((((((....)))))))).(((........)))..))))))))....... ( -42.90) >consensus UCUCUCGCCAUGAACCAGCUUACAAUCGCGAUUUUCUGGCUGGCAUGCAUUAACUUCGCCUCUUCGACUUGCUGCACUUCUGCGGCGACUCUGCACUUCAAGAUCCGUGGCGGAGUCUGU .(((.(((((((.....((........))((((((..(((.(((.............))).....((.((((((((....)))))))).)).)).)...))))))))))))))))..... (-31.51 = -31.70 + 0.19)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:38:39 2006