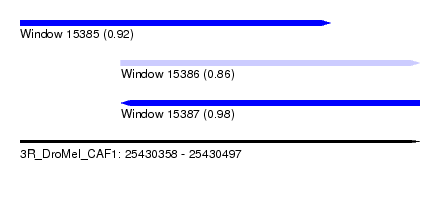
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 25,430,358 – 25,430,497 |
| Length | 139 |
| Max. P | 0.980843 |

| Location | 25,430,358 – 25,430,466 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 80.58 |
| Mean single sequence MFE | -26.42 |
| Consensus MFE | -13.88 |
| Energy contribution | -13.68 |
| Covariance contribution | -0.20 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.48 |
| Structure conservation index | 0.53 |
| SVM decision value | 1.15 |
| SVM RNA-class probability | 0.922881 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 25430358 108 + 27905053 AAUGUUAUCGUUGUAAAUUAUAAAAUGGAAUCCAUUAAUGCAGCCAGC-ACACAUACUAGAGUGUACAUACAUAGUGUAUAGU---GUGUGUGGGUGCAGCCGCACAGCGGA .......(((((((.........(((((...)))))..((((.(((.(-((((((.......(((((((.....)))))))))---)))))))).)))).....))))))). ( -33.81) >DroSec_CAF1 113184 93 + 1 AAUGUUAUCGUUGUAAAUUAUAAAAUGGAAUCCAUUAAUGCAGCCAGAUAUGCA----------CACAUACAUAGUG---------GUGUGUUGGUGCAGCCGCACAGUGGA .......(((((...........)))))..((((((..(((.((......((((----------(((((((......---------))))))..))))))).))).)))))) ( -21.40) >DroSim_CAF1 115211 86 + 1 AAUGUUAUCGUUGUAAAUUAUAAAAUGGAAUCCAUUAAUGCAAC-------ACA----------UACACACAUAGUG---------GUGUGUGGGUGCAGCCGCACAGCGGA .......(((((((.........(((((...)))))..((((.(-------(((----------(((.(((...)))---------)))))))..)))).....))))))). ( -26.10) >DroEre_CAF1 120419 99 + 1 AAUGUUAUCGUUGUAAAUUAUAAAAUGGAAUCCAUUAAUGCAGCCAGAUAUGCA----------CACAUGCAUAGUAUAUUGC---GUGCAUAGAUGCACCCACACAGCGGA .......(((((((.........(((((...)))))...((((.....((((((----------....)))))).....))))---(((((....)))))....))))))). ( -24.70) >DroYak_CAF1 123342 102 + 1 AAUGUUAUCGUUGUAAAUUAUAAAAUGGAAUCCAUUAAUGCAGCCAGAUGUGCA----------CACAUACAUAGUAUAUACUGUAGUGUGUAGAUGCACCCACACAGCGGA .......(((((((.........(((((...))))).............(((((----------((((((((((((....))))).)))))).).)))))....))))))). ( -26.10) >consensus AAUGUUAUCGUUGUAAAUUAUAAAAUGGAAUCCAUUAAUGCAGCCAGAUAUGCA__________CACAUACAUAGUGUAU______GUGUGUAGGUGCAGCCGCACAGCGGA .......(((((((.........(((((...)))))..((((((.......))..............((((((.............))))))...)))).....))))))). (-13.88 = -13.68 + -0.20)


| Location | 25,430,393 – 25,430,497 |
|---|---|
| Length | 104 |
| Sequences | 5 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 78.19 |
| Mean single sequence MFE | -28.72 |
| Consensus MFE | -15.96 |
| Energy contribution | -15.64 |
| Covariance contribution | -0.32 |
| Combinations/Pair | 1.38 |
| Mean z-score | -2.05 |
| Structure conservation index | 0.56 |
| SVM decision value | 0.81 |
| SVM RNA-class probability | 0.855351 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 25430393 104 + 27905053 UAAUGCAGCCAGC-ACACAUACUAGAGUGUACAUACAUAGUGUAUAGU---GUGUGUGGGUGCAGCCGCACAGCGGAAUGGUGGGCCAUCUAUGCAAAAUUCAUUUGC ...((((.(((.(-((((((.......(((((((.....)))))))))---)))))))).)))).((((...)))).((((....))))....(((((.....))))) ( -33.51) >DroSec_CAF1 113219 89 + 1 UAAUGCAGCCAGAUAUGCA----------CACAUACAUAGUG---------GUGUGUUGGUGCAGCCGCACAGUGGAAUGGCGGGCCAUCUAUGCAAAAUUCAUUUGC ...((((...((((.((((----------(((((((......---------))))))..))))).((((.((......))))))...)))).))))............ ( -26.60) >DroSim_CAF1 115246 82 + 1 UAAUGCAAC-------ACA----------UACACACAUAGUG---------GUGUGUGGGUGCAGCCGCACAGCGGAAUGGCGGGCCAUCUAUGCAAAAUUCAUUUGC ...((((.(-------(((----------(((.(((...)))---------)))))))..)))).((((.((......)))))).........(((((.....))))) ( -26.00) >DroEre_CAF1 120454 95 + 1 UAAUGCAGCCAGAUAUGCA----------CACAUGCAUAGUAUAUUGC---GUGCAUAGAUGCACCCACACAGCGGAAUGGCGGGCCAUCUAUGCAAAAUGCAAUUGC ....(((......((((((----------....))))))....(((((---(((((((((((..(((.(.((......))).))).))))))))))...))))))))) ( -30.80) >DroYak_CAF1 123377 98 + 1 UAAUGCAGCCAGAUGUGCA----------CACAUACAUAGUAUAUACUGUAGUGUGUAGAUGCACCCACACAGCGGAAUGGCGAGCCAUCUAUGCAAAAUUCAUUUGC ...((((((((...(((((----------((((((((((((....))))).)))))).).)))))((.......))..))))((....))..))))............ ( -26.70) >consensus UAAUGCAGCCAGAUAUGCA__________CACAUACAUAGUGUAU______GUGUGUAGGUGCAGCCGCACAGCGGAAUGGCGGGCCAUCUAUGCAAAAUUCAUUUGC ....((((........................((((...)))).........((((((((((...((((.((......))))))..))))))))))........)))) (-15.96 = -15.64 + -0.32)



| Location | 25,430,393 – 25,430,497 |
|---|---|
| Length | 104 |
| Sequences | 5 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 78.19 |
| Mean single sequence MFE | -29.24 |
| Consensus MFE | -15.92 |
| Energy contribution | -14.92 |
| Covariance contribution | -1.00 |
| Combinations/Pair | 1.32 |
| Mean z-score | -2.50 |
| Structure conservation index | 0.54 |
| SVM decision value | 1.87 |
| SVM RNA-class probability | 0.980843 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 25430393 104 - 27905053 GCAAAUGAAUUUUGCAUAGAUGGCCCACCAUUCCGCUGUGCGGCUGCACCCACACAC---ACUAUACACUAUGUAUGUACACUCUAGUAUGUGU-GCUGGCUGCAUUA (((((.....)))))...(((((....)))))((((...)))).((((.((((((((---(.(((((.....)))))(((......))))))))-).))).))))... ( -30.40) >DroSec_CAF1 113219 89 - 1 GCAAAUGAAUUUUGCAUAGAUGGCCCGCCAUUCCACUGUGCGGCUGCACCAACACAC---------CACUAUGUAUGUG----------UGCAUAUCUGGCUGCAUUA (((((.....)))))...(((((....))))).....(((((((((((((((((...---------.....))).)).)----------)))......)))))))).. ( -25.00) >DroSim_CAF1 115246 82 - 1 GCAAAUGAAUUUUGCAUAGAUGGCCCGCCAUUCCGCUGUGCGGCUGCACCCACACAC---------CACUAUGUGUGUA----------UGU-------GUUGCAUUA (((((.....)))))...(((((....))))).....(((((((.(((..((((((.---------.....))))))..----------)))-------))))))).. ( -27.10) >DroEre_CAF1 120454 95 - 1 GCAAUUGCAUUUUGCAUAGAUGGCCCGCCAUUCCGCUGUGUGGGUGCAUCUAUGCAC---GCAAUAUACUAUGCAUGUG----------UGCAUAUCUGGCUGCAUUA ((((((((....((((((((((((((((((......)).)))))).)))))))))).---)))))....((((((....----------))))))......))).... ( -37.10) >DroYak_CAF1 123377 98 - 1 GCAAAUGAAUUUUGCAUAGAUGGCUCGCCAUUCCGCUGUGUGGGUGCAUCUACACACUACAGUAUAUACUAUGUAUGUG----------UGCACAUCUGGCUGCAUUA (((((.....))))).......((..((((..((((...))))((((....(((((.((((..........))))))))----------)))))...)))).)).... ( -26.60) >consensus GCAAAUGAAUUUUGCAUAGAUGGCCCGCCAUUCCGCUGUGCGGCUGCACCCACACAC______AUACACUAUGUAUGUG__________UGCAUAUCUGGCUGCAUUA (((.........(((((((((((....)))).....((((.((......)).))))............)))))))...............((.......))))).... (-15.92 = -14.92 + -1.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:38:34 2006